Abstract
Download PDF
Full Article
Bacterial Diversity in Two Aerated Lagoons of a Pulp and Paper Effluent and their Interaction with a Commercial Inoculum using PCR-DGGE
Ana M. Bailón-Salas,a,δ Luis A. Ordaz-Díaz,b Sergio Valle-Cervantes,a Javier López-Miranda,a Norma Urtiz-Estrada,c Jesús B. Páez-Lerma,a Gerardo D. de León-Mata b and Juan A. Rojas-Contreras a,*
Aerated lagoons are a main unit operation for wastewater treatment in the paper industry. Many such operations involve inoculation with bacterial formulations in which in situ effectiveness has not been proven; this can be translated into low efficiency in treatment and unnecessary investments. Lack of knowledge of bacterial biodiversity present in a lagoon limits the capacity to exploit the maximum degradation. To overcome such problems, various methods to identify and study these microorganisms have been developed. In this study, a PCR-DGGE analysis was performed to estimate the bacterial diversity and to verify the presence of bacteria present in a commercial inoculum in two aerated lagoons of a pulp and paper effluent. Phylogenetic affiliation of predominant member’s correspondent to γ- and β-proteobacteria and Firmicutes were found. The dominant bacteria present in lagoon 2 belonged to the following genus Microbacterium sp. Rhodocyclaceae sp., Eubacterium sp. and B. subtilis. In lagoon 1 the dominant genus included Microbacterium sp., Rhodocyclaceae sp. Tepidimonas sp., Acetanaerobacterium sp., and Flavobacteria sp. The two characterized lagoons were not similar to the commercial inoculum. In addition, non-dominant bacteria (less relative intensity) were composed mostly of bacteria of the commercial inoculum.
Keywords: Pulp and paper effluent; Bacterial diversity; PCR-DGGE; 16S rDNA; Commercial inoculum
Contact information: a: Chemical and Biochemical Engineering Department, Durango Institute of Technology (ITD), Durango, México; b: Environmental Engineering Technology, Universidad Politécnica de Durango, Durango, México; c: Facultad de Ciencias Químicas, Universidad Juárez del Estado de Durango, Durango, México; δPhD Student; *Corresponding author: juanroco@hotmail.com
INTRODUCTION
Wastewaters of pulp-and-paper mills can be potentially polluting and very dangerous, so they should be treated before being discharged (Ashrafi et al. 2015). The most common aerobic biological methods used in the treatment of pulp mill effluents are aerated lagoons (Bajpai 2012). Of the microorganisms involved in the depuration process, bacteria stand out; they are able to convert organic matter to carbon dioxide, water, and biomass, which can be removed by physical methods (Welander et al. 1997; Forster et al. 2003). The stability and permanence of bacteria in the system ensures an efficient process. However, lagoons are subjected to various perturbations such as variations in pH, high organic loads, presence of toxic compounds, and seasonal changes (Mueller et al. 1977). To counteract these effects, a biomass support material (Welander et al. 1997) or lagoons that are inoculated with commercial inoculum are introduced.
The application of traditional and molecular techniques has provided a glimpse into the industry’s “black box” and has given information to improve the wastewater treatment process. Currently, a great variety of molecular methods are available that provide more information than the widely known culture methods. Fingerprint methods, such as DGGE, can be used to monitor through the purification process (Bailón-Salas et al. 2017). However, few reports concerning the study of bacterial communities in aerated lagoons in a pulp and paper effluent exist (Liss and Allen 1992; Yu and Mohn 2001). The lack of knowledge of the bacterial communities present in the aerated lagoons of the pulp and paper effluent limits the capacity to take advantage of maximizing its purifying capacity.
The wastewater treatment plant (WWTP) of this study is located in northern Mexico and receives effluent from the pulp and paper mill industry. This plant performs a pre-treatment based on sieves, using a clarifier as the primary treatment, a secondary treatment based on two lagoons in series, posteriorly a packaged filter, and a chlorine-based final disinfection process. This study was performed in the secondary treatment. The lagoons are inoculated daily with a commercial inoculum, whose in situ effectiveness has not been proven.
The aim of this research is to verify the presence of bacteria from the commercial inoculum and identify the bacterial community composition in two aerated lagoons of pulp and paper effluent using PCR-DGGE. A 16S rDNA-based approach to determine the bacterial community associated with commercial inoculum was used.
EXPERIMENTAL
Materials
Wastewater samples were collected from the two aerated lagoons of pulp and paper mill effluent from the WWTP located in northern Mexico. Dimensions of the lagoons are 20 m x 120 m x 7 m and an occupied volume of 93,480 m3. Samples were stored at 4 °C until used for DNA extraction and physicochemical analysis.
Temperature, dissolved oxygen (DO), electrical conductivity (EC), and pH were determined in situ with an HQ40d Portable device (HACH Company, Loveland CO, USA) for each lagoon. Settleable solids (SS) was analyzed gravimetrically. The biochemical oxygen demand (BOD5) was determined by a manometric-respirometric method with a BODTrak™ II apparatus (Hach Company, 2010, Loveland, Colorado). The measurement of chemical oxygen demand (COD) was determined using the closed reflux (colorimetric) method described in section 5220-D and turbidity was determined by a spectrophotometric method to analyze water and wastewater (Rice et al. 2012). Using a photometer HI83225 (HANNA Instruments Inc., Woonsocket, RI, USA) phosphorus, potassium, nitrates, and sulfates were determined.
Commercial inoculum preparation
The commercial culture of microorganisms (unknown composition) was cultured in 3 mL of Lysogeny Broth (LB) medium and incubated overnight at 37 °C. To verify which species are not present under different conditions (lagoon conditions), standard conditions based on Kumar et al. (2008) were used. The commercial formulation in potato dextrose agar (PDA) was grown to confirm or discard the hypothesis that the formulation is also composed of fungi.
DNA extraction
To compare the banding profiles, simple and composite samples (all other sampling points) of each lagoon and commercial inoculum were processed. To eliminate suspended particles, 40 mL of water samples were pre-filtered, centrifuged, and washed with a 2 mL buffer Tris-EDTA (TE). Following the manufacturer´s instructions, the total metagenomics DNA, using a soil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), was extracted. The extraction was visualized on 1% agarose gels stained with ethidium bromide (Bio-Rad Laboratories, Inc., Irvine, CA, USA).
Methods
Polymerase chain reaction (PCR) amplification and denaturing gradient gel electrophoresis (DGGE)
Bacterial DNA was amplified using 357f/518r primers (357f: 5-CGCCCGCCG-CGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3; 518r: 5-ATTACCGCGGCTGCTGG-3) (Lopez et al. 2003; Sánchez et al. 2007), which amplify the variable region V3 of the 16S rRNA gene. Following the manufacturer’s instructions, the PCR reactions were performed in 25-μL volumes using Taq DNA polymerase (PROMEGA, Madison, USA). The amplification reactions were performed in a thermal cycler (T100 of BioRad, Irvine, CA, USA) using the following cycling program: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s, and extension at 72 °C for 90 s. The final extension was at 65 °C for 10 min. All amplicons were analyzed on 1% agarose gels to confirm the size and yield; the bands were visualized in a PhotoDoc-It™ Imaging System (UVP LLC, Inc., Upland, CA, USA).
The DCode Universal Mutation Detection System (Bio-Rad Laboratories, Irvine, CA, USA) for the DGGE analysis was used. The PCR products (900 ng) were applied to polyacrylamide gel 8% (w/v) acrylamide/bisacrylamide, containing a 30% to 60% urea-formamide denaturing gradient. The gels were run with 1X TAE buffer at a voltage of 20 V for 10 min, 200 V for 120 min, and 120 V for 120 min at 60 °C (Lopez et al. 2003). For a higher resolution, the gel was silver-stained using the protocol described by Radojkovic and Kušic (2000). The bands were excised, crushed, and eluted in 30 µL of milliQ water. One microliter of eluted DNA for re-amplification was used, as described above. The PCR products with Wizard SV were purified gel and PCR Clean-Up System (Promega Corporation, Fitchburg, WI, USA); and sequenced by DNA Synthesis and the Sequencing Unit of the Institute of Biotechnology (UNAM, Morelos, Mexico).
Phylogenetic analysis
The sequences were compared with 16S rRNA gene sequences published in the National Center for Biotechnology Information DNA database using Basic Local Alignment Search Tool (BLAST) (Bethesda, MD, USA) and were aligned using ClustalW of BioEdit 7.2.5 version (Hall 1999). The phylogenetic relationships were constructed with the Molecular Evolutionary Genetics Analysis (MEGA) version 7.0 (Kumar et al. 2016) using the Neighbor-joining similarity coefficient.
Nucleotide sequence accession numbers
The sequences were deposited in the GenBank public database under the accession numbers: KY847917, KY847918, KY847919, KY847920, KY847921, KY847922, KY847923, KY847924, KY847925, KY847926, KY847927, KY847928, KY847929, KY847930, KY847931, KY847932, KY847933, and KY847934.
Analysis of DGGE profiles
The gel image was analyzed using Gelquant® software (Biochem, San Francisco, CA, USA). The DGGE profiles based on relative intensity (RI) of bands were analyzed and a presence-absence matrix was generated. The Shannon diversity index (H′), Shannon equitability index (EH′), richness (S) (total number of DGGE bands), and analysis of correlation (between lagoons and formulation) using BioDiversity Pro® version 2.0 software (Scottish Association for Marine Science and the Natural History Museum of London, London, UK) were calculated in each sample (Shannon 1948). The distance matrix for constructing a similitude dendrogram was also used.
RESULTS AND DISCUSSION
The samples were taken in March 2016 at 18 points for each lagoon (surface and 3 m of depth) (Fig. 1), and resulted in 36 samples (18 samples per lagoon).
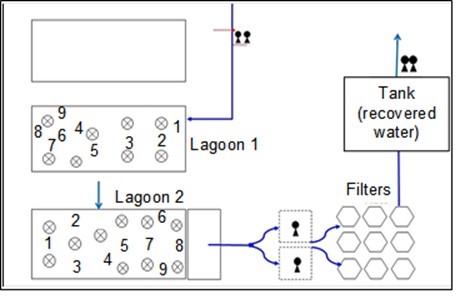
Fig. 1. Sampling sites along the lagoons
Physical and Chemical Characterization
Influent physicochemical characterizations during the sampling are shown in Table 1. The temperature in lagoons 1 and 2 was 20 °C ± 0.32 and 18.4 °C ± 0.62, respectively. A lower temperature in the lagoon 2 would have a selective effect on the community. The succession was also partly autogenic, a consequence of the community’s activities. For example, the reduction of degradable organic carbon depleted certain substrates, including methanol and resin acids. The DO in both lagoons was lower than 1.07 mg/L. The degradation of organic carbon depleted DO to very low levels at the beginning of the system. A slight increase in pH was observed in lagoon 2 (7.44 to 7.81). The BOD5 was higher in lagoon 1, because lagoon 1 was receiving the effluent and therefore was the lagoon with the highest organic load (Table 1); this value was close to the BOD-initial that was reported by Ordaz-Díaz et al. (2014) and Welander et al. (1997). In the case of sulfates, these are used by bacteria as an oxidizing agent, which is reduced to hydrogen sulfide, the gas responsible for bad odors. The changing physicochemical conditions in the lagoon were both a cause and a result of the community succession. The observed spatial gradients were due to the plug-flow regime of the lagoon and were generally gradual and constant. The characteristic pH for bleached kraft mill (BKM) effluents was reported by Schnell et al. (2000). The turbidity in lagoon 1 was higher, which again is consistent with the expected greater organic load. In addition, the free bacteria are too small to sediment, giving a characteristic turbidity to the effluent. Pulp mill effluents have a low nitrogen content, because of the low C:N ratio of wood. Nitrogen fixation has been reported in aerated lagoons treating pulp mill effluents (Bruce and Clark 1994; Clark et al. 1997). Certain physicochemical factors appeared to be particularly important in effecting community changes.
Table 1. Physicochemical Characterization of Water Samples Collected in Both Aerated Lagoons

DGGE Analysis of Bacterial Community
Simple samples (a specific point) and composites (all other points except the point corresponding to the simple sample) of each lagoon were analyzed to verify the homogeneity of the points sampled. Thus, the totals of samples analyzed in both lagoons were 4 (2 per lagoon). The simple and composite sample of lagoon 1 was designated as L1 (sampling point 6) and L1C, respectively. Likewise, in lagoon 2 the samples were denominate as L2 (sampling point 3) and L2C. DNA fragment of the commercial inoculum were designated as F, (obtained from LB medium). Amplicons were also loaded onto the DGGE gel that corresponded to previously identified isolates, to favor the process of identification of bands.
A DGGE profile was used to investigate the bacterial community changes, presence of the commercial inoculum, and homogeneity of sampled points in pulp and paper aerated-lagoons. There were 32 band positions detected in the denaturing gradient gels (Fig. 2).
A matrix was constructed based on the presence or absence of bands (based relative intensity of each DGGE band). The percentages of abundance of bands detected in DGGE gel are shown in Table 2. The species richness was as follows: 14 in the formulation lane (F), 21 in L2 and L2C, and 19 and 17 in L1 and L1C, respectively. The highest species richness was presented in lagoon 2 (21 species). This may have been due to the bacteria-leaving lagoon 1 along with the effluent. The bacteria present in the formulation were not part of the dominant bacteria in both lagoons. In addition, non-dominant bacteria (less relative intensity) were composed mostly of bacteria of the commercial inoculum.
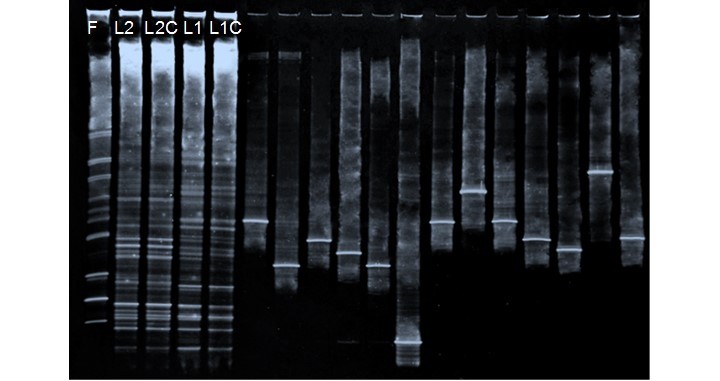
Fig. 2. DGGE-profiles of metagenomics DNA extracted from commercial formulation and water collected in two lagoons
Table 2. Percentage of Abundance of Bacterial Communities Associated with Commercial Inoculum and Two Aerated Lagoons
The measured diversity in the five samples showed ranging high levels. The Shannon index (H ‘) showed an equitable abundance distribution: formulation = 1.039, L2 = 1.233, L2C = 1.207, L1 = 1.172 and L1C = 1.088 (Table 2). Shannon’s equitability assumed a value between 0 and 1, with 1 being complete evenness.
Table 3. DGGE Shannon Diversity Index and Shannon Equitability Index to Commercial Inoculum and the Two Aerated Lagoons
The similarity analysis showed a relationship between L2 and L2C (89.2 and 94.31, respectively) and between L1 and L1C (83.4 and 93.4, respectively). However, no significant correlation was shown between the formulation and both gaps (Fig. 3).
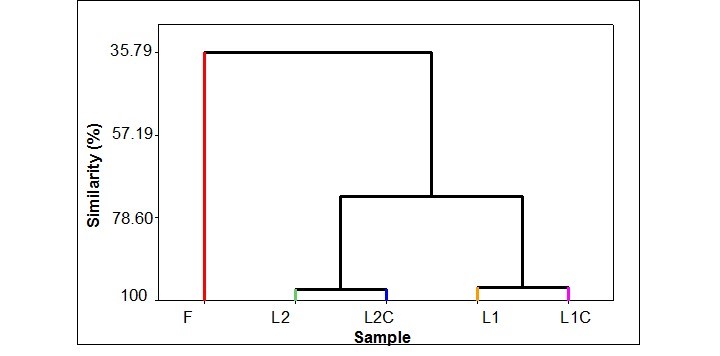
Fig. 3. Dendrogram (single link) of correlation coefficient distance
Sequencing of DGGE Bands and Phylogenetic Analysis
The phylogenetic affiliation of predominant members appertained for γ- and β- proteobacteria and Firmicutes. Each sequence submitted to the BLAST search is shown in the Table 4 and Fig. 4.
Phylogenetic identification of the dominant bacteria in lagoon 2 belonged to the genus Microbacterium sp. Rhodocyclaceae sp., Eubacterium sp. and B. subtilis specie. In lagoon 1 corresponded to the dominant genus Microbacterium sp., Rhodocyclaceae sp. Tepidimonas sp., Acetanaerobacterium sp., and Flavobacteria sp.
Both Microbacterium sp. and Rhodocyclaceae sp. were present in both lagoons as dominant bacterial. These two bacteria are related to polycyclic aromatic hydrocarbons (PAH) degradation (Regonne et al. 2013). Chalbot et al. (2006) reported the presence of PAHs in recycled paper processing operations. Furthermore, it has been reported by Jung et al. (2002) that Microbacterium is able to degrade lignin; Tripathi et al. (2014) studied the production of lipases (isolated from residual sludge from a paper effluent); and Sun et al. (2007) studied the production of chitosanase by this bacterium. Chitosan is used as a coagulant in the treatment of paper effluents (Pokhrel and Viraraghavan 2004).
Table 4. Phylogenetic Sequence Affiliation and Similarity from DGGE gels
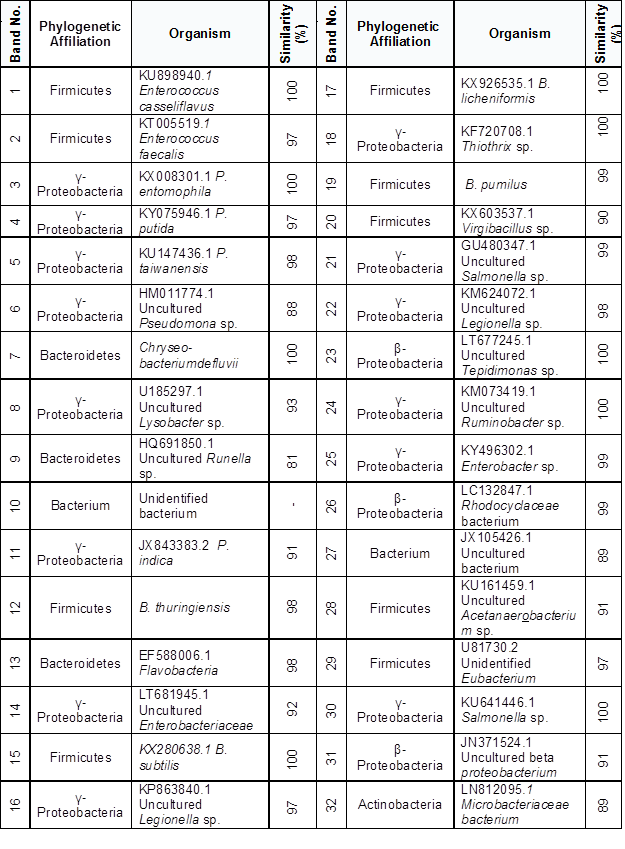
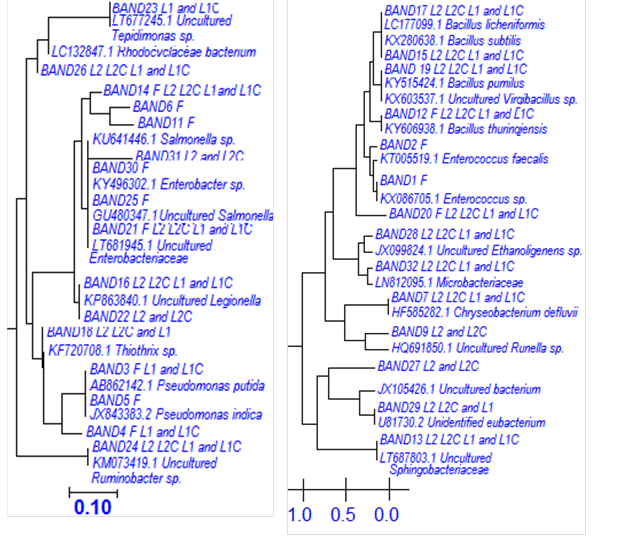
Fig. 4. Neighbor-joining phylogenetic tree based on partial 16S rDNA sequences derived from DGGE bands affiliated to γ- and β-Proteobacteria and Firmicutes
The moderately dominant bacteria in lagoon 1 belong to C. defluvii, B. thuringiensis (present in the commercial inoculum), Uncultured Enterobacteriaceae, B. subtilis, Uncultured Legionella sp., B. licheniformis, Thiothrix sp., and Uncultured Ruminobacter sp. In lagoon 2 they belong to Uncultured Runella sp., B. licheniformis, Thiothrix sp., Uncultured Legionella sp., Uncultured bacterium, Uncultured Acetanaerobacterium sp., Flavobacteria sp., and Uncultured Legionella sp.
The non-dominant bacteria in lagoon 1 correspond to Pseudomonas entomophila, P. putida, B. pumilus, Virgibacillus sp., and Uncultured Salmonella sp. And in lagoon 2 they belong to C. defluvii, B. thuringiensis, Uncultured Enterobacteriaceae, B. pumilus, Virgibacillus sp., Uncultured Salmonella sp., Ruminobacter sp., and Microbacteriaceae bacterium.
Pulp and paper mills are an important source for the isolation of microorganisms and for the production of enzymes by biotechnological methods, such as those identified in this work. Such is the case of B. pumilus and B. megaterium, in which studies were conducted to produce thermostable xylanases for pulp bleaching and to degrade hemicellulose, respectively (Sindhu et al. 2006; Battan et al. 2007). Kaushal et al. (2015) studied production of cellulase-free endo-beta-1,4-xylanase of B. subtilis isolated from paper mill effluent. Wongwilaiwalin et al. (2013) studied capability on the degradation of agricultural residues and industrial pulp waste with CMCase, xylanase, and β- glucanase activities of Acetanaerobacterium sp. in coexistence with ligninolytic Ureibacillus sp. showed the highest power of hydrolysis. Microbial β-mannanases (mannan-degrading enzymes, main constituent of the hemicellulose fraction) from B. subtilis (Jiang et al. 2006; Chauhan et al. 2012), Chryseobacterium sp. (Chauhan et al. 2012) and B. licheniformis (Songsiriritthigul et al. 2010; Ge et al. 2016) have been characterized.
Furthermore, the genus Bacillus has been extensively identified in biological effluents from pulp and paper mills (Bailón-Salas et al. 2017). B. subtilis has also been isolated from different gastro-intestinal tract regions of snails, showing high Avicelase activity (Dar et al. 2015). Raj et al. (2007) isolated, from activated sludge from pulp and paper mill effluent to B. thuringiensis, a ligninolytic bacterium that reduces the color and kraft-lignin. B. megaterium and B. thuringiensis used 2,3,4,6-tetrachlorophenol as a source of carbon and energy and were capable of degrading this compound (Karn et al. 2014). B. pumilus and Virgibacillus sp. have been characterized as Cr-resistant bacteria, reducing COD and Cr; the bacteria were isolated from the rhizosphere of Spirodela polyrrhiza (Singh et al. 2016) and in the sediments of a mangrove wetland (Liu et al. 2017).
The genus Chryseobacterium was reported by Chiellini et al. (2014) in the water circuit process of the paper mill, and was characterized as a plant-growth-promoting bacterium (Montero-Calasanz et al. 2013). Enterococcus casseliflavus, Microbacterium, Flavobacterias, Enterobacter sp., Chryseobacterium sp., Tepidimonas sp., and Pseudomonas sp., were isolated from untreated paper mill pulps (Suihko and Shooter 2009).
A pentachlorophenol degrading bacterium Flavobacterium (Gonzalez and Hu 1991; Suihko and Shooter 2009) and Tepidimonas (Tiirola et al. 2009) have been reported in the process of paper production. P. putida and Enterobacter sp. reduced color, BOD, COD, and phenolic compounds in a pulp and paper mill effluent and sulfide and heavy metals were removed (Chandra 2001). The Enterobacteriaceae family reduce lignin and color (Singh et al. 2011).
Thiothrix sp. in pulp and paper activated sludge (Dumonceaux et al. 2006) has been characterized. This bacterium causes filamentous bulking, giving settling problems in secondary clarifiers (Pelletier et al. 2007). In contrast, taking advantage of the native bacterial flora to treat the effluent is a viable option. Ordaz-Diaz et al. (2014) generated an isolated bacterial formulation from wastewater from lagoon 1. The efficiency was measured in the BODTrack system; this bacterial formulation exceeded the efficiency of the commercial inoculum. A mixed culture of B. subtilis and Klebsiella pneumonia resulted in a better decolorization efficiency and reduction of pollution parameters (COD and BOD) of paper mill effluent (Yadav and Chandra 2015).
The PCR–DGGE- was demonstrated to be a dual tool that applied the numerical analysis to the results obtained in the techniques of molecular identification.
CONCLUSION
- The bacteria present in the formulation were not part of the dominant bacteria in both lagoons. In addition, non-dominant bacteria (less relative intensity) were composed mostly of bacteria of the commercial formulation.
- The results indicated that the greatest richness of species was present in lagoon 2 (21). The distribution of relative abundance achieved was equitable (uniform) in both lagoons. The similarity matrix showed a considerable relationship between L2 and L2C, as well as between L1 and L1C.
- The available evidence supported the conclusion that the community’s spatial succession in the lagoons involved an increased phylogenetic diversity from the beginning to the end of the lagoons.
- The PCR-DGGE method applied in this study has allowed the authors to verify whether bacteria that make up a commercial inoculum form part of the active microbiota in two aerated lagoons of pulp and paper effluent.
ACKNOWLEDGMENTS
The support of the Science and Technology National Council (CONACyT) was gratefully appreciated, as it was received by one of the writers for the scholarship (419668/257494).
REFERENCES CITED
Ashrafi, O., Yerushalmi, L., and Haghighat, F. (2015). “Wastewater treatment in the pulp-and-paper industry: A review of treatment processes and the associated greenhouse gas emission,” Journal of Environmental Management 158, 146-157. DOI: 10.1016/j.jenvman.2015.05.010
Bailón-Salas, A. M., Medrano-Roldán, H., Valle-Cervantes, S., Ordaz-Díaz, L. A., Urtiz-Estrada, N., and Rojas-Contreras, J. A. (2017). “Review of molecular techniques for the identification of bacterial communities in biological effluent treatment facilities at pulp and paper mills,” BioResources (Submitted).
Bajpai, P. (2012). “Biological treatment of pulp and paper mill effluents,” in: Biotechnology for Pulp and Paper Processing, Springer US, Boston, MA, pp. 211-261. DOI: 10.1007/978-1-4614-1409-4_1
Battan, B., Sharma, J., Dhiman, S. S., and Kuhad, R. C. (2007). “Enhanced production of cellulase-free thermostable xylanase by Bacillus pumilus ASH and its potential application in paper industry,” Enzyme and Microbial Technology, 41(6), 733-739. DOI: 10.1016/j.enzmictec.2007.06.006
Bruce, M. E., and Clark, T. A. (1994). “Klebsiella and nitrogen fixation in pulp and paper mill effluents and treatment systems,” Appita Journal 47(3), 231-237.
Chalbot, M. C., Vei, I., Lykoudis, S., and Kavouras, I. G. (2006). “Particulate polycyclic aromatic hydrocarbons and n-alkanes in recycled paper processing operations,” Journal of Hazardous Materials 137(2), 742-751. DOI: 10.1016/j.jhazmat.2006.04.003.
Chandra, R. (2001) “Microbial decolourisation of pulp and paper mill effluent in presence of nitrogen and phosphorus by activated sludge process,” J. Environ. Biol. 2001 Jan; 22(1) 23-27. PMID: 11480347
Chauhan, P. S., Puri, N., Sharma, P., and Gupta, N. (2012). “Mannanases: Microbial sources, production, properties and potential biotechnological applications,” Appl. Microbiol. Biotechnol. 93, 1817. DOI: 10.1007/s00253-012-3887-5
Chiellini, C., Iannelli, R., Lena, R., Gullo, M., and Petroni, G. (2014). “Bacterial community characterization in paper mill white water,” BioResources 9(2), 2541-2559. DOI: 10.15376/biores.9.2.2541-2559
Clark, T. A., Dare, P. H., and Bruce, M. E. (1997). “Nitrogen fixation in an aerated stabilization basin treating bleached kraft mill wastewater,” Water Environment Research 69(5), 1039-1046. DOI: 10.2175/106143097X125740
Dar, M. A., Pawar, K. D., Jadhav, J. P., and Pandit, R. S. (2015). “Isolation of cellulolytic bacteria from the gastro-intestinal tract of Achatina fulica (Gastropoda: Pulmonata) and their evaluation for cellulose biodegradation,” International Biodeterioration & Biodegradation 98, 73-80. DOI: 10.1016/j.ibiod.2014.11.016
Dumonceaux, T. J., Hill, J. E., Pelletier, C. P., Paice, M. G., Van Kessel, A. G., and Hemmingsen, S. M. (2006). “Molecular characterization of microbial communities in Canadian pulp and paper activated sludge and quantification of a novel Thiothrix eikelboomii-like bulking filament,” Canadian Journal of Microbiology 52(5), 494-500. DOI: 10.1139/W05-160
Forster, S., Lappin-Scott, H. M., Snape, J. R., and Porter, J. (2003). “Rains, drains and active strains: Towards online assessment of wastewater bacterial communities,” Journal of Microbiological Methods 55(3), 859-864. DOI: 10.1016/j.mimet.2003.08.004
Ge, J., Du, R., Zhao, D., Song, G., Jin, M., and Ping, W. (2016). “Kinetic study of a β-mannanase from the Bacillus licheniformis HDYM-04 and its decolorization ability of twenty-two structurally different dyes,” SpringerPlus 5(1), 1824. DOI: 10.1186/s40064-016-3496-3
Gonzalez, J. F. and Hu, W.S. (1991). “Effect of glutamate on the degradation of pentachlorophenol by Flavobacterium sp.,” Appl Microbiol Biotechnol 35, 100-104. DOI: 10.1007/BF00180644
Hall, A. (1999). “BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT,” in: Nucleic Acids Symposium Series 41, 95-98.
Jiang, Z., Wei, Y., Li, D., Li, L., Chai, P. P., and Kusakabe, I. (2006). “High-level production, purification and characterization of a thermostable β-mannanase from the newly isolated Bacillus subtilis WY34,” Carbohydrate Polymers 66, 88-96. DOI: 0.1016/j.carbpol.2006.02.030
Jung, S., Kim H., Koo Y., Oh H., Park C., and Yoon B. (2002). “Microorganism Microbacterium sp. pa5 (kctc 10172bp) capable of degrading lignin,” Korean Patent No. KR20030071077.
Karn, S. K., Reddy, M. S., and Chakrabarti, S. K. (2014). “Biomineralization of 2, 3, 4, 6-tetrachlorophenol by Bacillus sp. and Staphylococcus sp. isolated from secondary sludge of pulp and paper mill,” Bioremediation Journal 18(2), 93-99. DOI: 10.1080/10889868.2014.889073
Kaushal, R., Sharma, N., and Dogra, V. (2015). “Optimization of the production and molecular characterization of cellulase-free xylanase from an alkalophillic Bacillus subtilis SD8 isolated from paper mill effluent,” Appl. Biochem. Microbiol. 51, 551. DOI: 10.1134/S0003683815050117
Kumar, R., Rastogi, S., and Kumar, A. (2008). “Microbial consortium useful as seeding material for BOD analysis of pulp and paper industrial wastewater,” U.S. Patent No. 7,429,478, 30 Sept. 2008.
Kumar, S., Stecher, G., and Tamura, K. (2016). “MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets,” Molecular Biology and Evolution msw054. DOI: 10.1093/molbev/msw054
Liss, S. N., and Allen, D. G. (1992). “Microbiological study of bleached kraft pulp mill aerated lagoon,” Journal of Pulp and Paper Science 18(6), 216-221.
Liu, M., Cui, Y., Chen, Y. Q., Lin, X., Huang, H. Q., and Bao, S. (2017). “Diversity of Bacillus-like bacterial community in the sediments of the Bamenwan mangrove wetland in Hainan, China,” Canadian Journal of Microbiology 63(3), 238-245. DOI: 10.1139/cjm-2016-0449
Lopez, I., Ruiz-Larrea, F., Cocolin, L., Orr, E., Phister, T., Marshall, M., VanderGheynst, J., and Mills, D. A. (2003). “Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis,” Applied and Environmental Microbiology 69(11), 6801-6807. DOI: 10.1128/AEM.69.11.6801-6807.2003
Montero-Calasanz, M. C., Göker, M., Rohde, M., Spröer, C., Schumann, P., Busse, H. J., et al. (2013). “Chryseobacterium hispalense sp. nov., a plant-growth-promoting bacterium isolated from a rainwater pond in an olive plant nursery, and emended descriptions of Chryseobacterium defluvii, Chryseobacterium indologenes, Chryseobacterium wanjuense and Chryseobacterium gregarium,” International Journal of Systematic and Evolutionary Microbiology 63(12), 4386-4395. DOI: 10.1099/ijs.0.052456-0
Mueller, J. C., Leach, J. M., and Walden, C. C. (1977). “Detoxification of bleached kraft mill effluents- A manageable problem,” in: TAPPI Environmental Conference, Chicago, IL, pp. 25-27.
Ordaz-Díaz, L. A., Rojas-Contreras, J. A., Rutiaga-Quiñones, O. M., Moreno-Jiménez, M. R., Alatriste-Mondragón, F., and Valle-Cervantes, S. (2014). “Microorganism degradation efficiency in BOD analysis formulating a specific microbial consortium in a pulp and paper mill effluent,” BioResources 9(4), 7189-7197. DOI: 10.15376/biores.9.4.7189-7197
Pelletier, C., Fitzsimmons, M. A., Deschênes, S., and Paice, M. (2007). “Impact of septic compounds and operational conditions on the microbiology of an activated sludge system,” Water Science and Technology 55(6), 135-142. DOI: 10.2166/wst.2007.221
Pokhrel, D., and Viraraghavan, T. (2004). “Treatment of pulp and paper mill
Wastewater- A review,” Science of the Total Environment 333(1-3), 37-58. DOI: 10.1016/j.scitotenv.2004.05.017
Radojkovic, D., and Kušic, J. (2000). “Silver staining of denaturing gradient gel electrophoresis gels,” Clinical Chemistry 46(6), 883-884.
Raj, A., Reddy, M. K., Chandra, R. Purohit, H. J., and Kapley, A. (2007). “Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill,” Biodegradation, 18(6), 783-792. DOI: 10.1007/s10532-007-9107-9
Regonne, R. K., Martin, F., Mbawala, A., Ngassoum, M. B., and Jouanneau, Y. (2013). “Identification of soil bacteria able to degrade phenanthrene bound to a hydrophobic sorbent in situ,” Environmental Pollution 180, 145-151. DOI: 10.1016/j.envpol.2013.04.038
Rice, E. W., Baird, R. B., Eaton, A. D., and Clesceri, L. S. (2012). Standard Methods for the Examination of Water and Wastewater (22nd Ed.), American Public Health Association, New York.
Sánchez, O., Gasol, J. M., Massana, R., Mas, J., and Pedrós-Alió, C. (2007). “Comparison of different denaturing gradient gel electrophoresis primer sets for the study of marine bacterioplankton communities,” Applied and Environmental Microbiology 73(18), 5962-5967. DOI: 10.1128/AEM.00817-07
Schnell, A., Steel, P., Melcer, H., Hodson, P. V., and Carey, J. H. (2000). “Enhanced biological treatment of bleached kraft mill effluents – I. Removal of chlorinated organic compounds and toxicity,” Water Research 34(2), 493-500. DOI: 10.1016/S0043-1354(99)00160-8
Shannon, C. E. (1948). “A mathematical theory of communication,” The Bell System Technical Journal 27(3), 379-423. DOI: 10.1002/j.1538-7305.1948.tb01338.x
Sindhu, I., Chhibber, S., Capalash, N., and Sharma, P. (2006). “Production of cellulase-free xylanase from Bacillus megaterium by solid state fermentation for biobleaching of pulp,” Current Microbiology 53(2), 167-172. DOI: 10.1007/s00284-006-0051-4
Singh, A., Vyas, D., and Malaviya, P. (2016). “Two-stage phyto-microremediation of tannery effluent by Spirodela polyrrhiza (L.) Schleid and chromium resistant bacteria,” Bioresource Technology 216, 883-893. DOI: 10.1016/j.biortech.2016.06.025
Singh, Y. P., Dhall, P., Mathur, R. M., Jain, R. K., Thakur, V. V., Kumar, et al. (2011). “Bioremediation of pulp and paper mill effluent by tannic acid degrading Enterobacter sp.,” Water, Air, & Soil Pollution 218(1-4), 693-701. DOI: 10.1007/s11270-010-0678-4
Songsiriritthigul, C., Buranabanyat, B., Haltrich, D., and Yamabhai, M. (2010). “Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1, 4-β-mannosidase from Bacillus licheniformis in Escherichia coli,” Microbial Cell Factories 9(1), 20. DOI: 10.1186/1475-2859-9-20
Suihko, M. L., and Shooter, E. (2009). “Characterisation of aerobically grown non-spore-forming bacteria from paper mill pulps containing recycled fibres,” J. Ind. Microbiol. Biotechnol. 36, 53. DOI: 10.1007/s10295-008-0472-0
Sun, Y., Han, B., Liu, W., Zhang, J., and Gao, X. (2007). “Substrate induction and statistical optimization for the production of chitosanase from Microbacterium sp. OU01,” Bioresource Technology 98(8), 1548-1553. DOI: 10.1016/j.biortech.2006.07.020
Tiirola, M., Lahtinen, T., Vuento, M., and Oker-Blom, C. (2009). “Early succession of bacterial biofilms in paper machines,” Journal of Industrial Microbiology & Biotechnology 36(7), 929-937.
Tripathi, R., Singh, J., Kumar Bharti, R., and Thakur, I. S. (2014). “Isolation, purification and characterization of lipase from Microbacterium sp. and its application in biodiesel production,” Energy Procedia 54, 518-529. DOI: 10.1016/j.egypro.2014.07.293
Welander, T., Löfgvist, A., and Selmer, A. (1997). “Upgrading aerated lagoons at pulp and paper mills,” Water Science and Technology 35(2-3), 117-122. DOI: 10.1016/S0273-1223(96)00922-5
Wongwilaiwalin, S., Laothanachareon, T., Mhuantong, W. Tangphatsornruang, S., Eurwilaichitr, L., Igarashi, Y., and Champreda, V. (2013). “Comparative metagenomic analysis of microcosm structures and lignocellulolytic enzyme systems of symbiotic biomass-degrading consortia,” Appl Microbiol Biotechnol 97: 8941. DOI:10.1007/s00253-013-4699-y
Yadav, S., and Chandra, R. (2015). “Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry,” Journal of Environmental Sciences 33, 229-238. DOI: 10.1016/j.jes.2015.01.018
Yu, Z., and Mohn, W. W. (2001). “Bacterial diversity and community structure in an aerated lagoon revealed by ribosomal intergenic spacer analyses and 16s ribosomal DNA sequencing,” Applied Environmental Microbiology 67(4), 1565-1574. DOI: 10.1128/AEM.67.4.1565-1574.2001
Article submitted: March 6, 2017; Peer review completed: May 25, 2017; Revised version received and accepted: June 2, 2017; Published: June 14, 2017.
DOI: 10.15376/biores.12.3.5487-5501
