Abstract
Symbiotic bacteria in the termite gut system may play an important role in lignin degradation that can assist the subsequent saccharification process. Pseudocitrobacter anthropi MP-4, which is capable of degrading lignin components and rapidly growing on various lignin analogue dyes, was successfully screened from the gut of a wood-feeding termite Microtermes pakistanicus. Further decolorization tests with this strain showed that the strain MP-4 potentially produced some relevant extracellular enzymes to participate in lignin degradation. The removal rate of chemical oxygen demand by this strain was recorded as high as 52.1% when it was incubated in a mineral-salt medium with lignin as the sole carbon source. For the degrading process of MP-4 on lignin, it was proposed through a series of evaluations by field emission scanning electron microscopy, Fourier transform infrared spectroscopy, thermogravimetric analysis, and pyrolysis gas chromatography/mass spectroscopy, that the lignin degradation mechanism of the strain MP-4 would primarily include the cleavage of various chemical linkages and the demethylation reactions. This resulted in a change in the S/G ratio and the disappearance of the biphenyl structure in the lignin components. Thus, these findings suggested that the strain MP-4 uniquely presented an attractive capability to deconstruct lignin components from biomass, which may be potentially valuable for a future industrial exploration.
Download PDF
Full Article
Biodegradation of Lignin via Pseudocitrobacter anthropi MP-4 Isolated From the Gut of Wood-feeding Termite Microtermes pakistanicus (Isoptera: termitidae)
Feng Li, Rongrong Xie, Nian Liang, Jianzhong Sun,* and Daochen Zhu *
Symbiotic bacteria in the termite gut system may play an important role in lignin degradation that can assist the subsequent saccharification process. Pseudocitrobacter anthropi MP-4, which is capable of degrading lignin components and rapidly growing on various lignin analogue dyes, was successfully screened from the gut of a wood-feeding termite Microtermes pakistanicus. Further decolorization tests with this strain showed that the strain MP-4 potentially produced some relevant extracellular enzymes to participate in lignin degradation. The removal rate of chemical oxygen demand by this strain was recorded as high as 52.1% when it was incubated in a mineral-salt medium with lignin as the sole carbon source. For the degrading process of MP-4 on lignin, it was proposed through a series of evaluations by field emission scanning electron microscopy, Fourier transform infrared spectroscopy, thermogravimetric analysis, and pyrolysis gas chromatography/mass spectroscopy, that the lignin degradation mechanism of the strain MP-4 would primarily include the cleavage of various chemical linkages and the demethylation reactions. This resulted in a change in the S/G ratio and the disappearance of the biphenyl structure in the lignin components. Thus, these findings suggested that the strain MP-4 uniquely presented an attractive capability to deconstruct lignin components from biomass, which may be potentially valuable for a future industrial exploration.
Keywords: Lignin degradation; Termite gut symbionts; Pseudocitrobacter anthropi; Microtermes pakistanicus
Contact information: Biofuels Institute of Jiangsu University, School of Environmental and Safety Engineering, Jiangsu University, No. 301 Xuefu Road, Jingkou District, Zhenjiang 212013, Jiangsu Province, China; *Corresponding authors: jzsun1002@ujs.edu.cn; dczhucn@hotmail.com
INTRODUCTION
Lignin is considered one of the most abundant renewable and natural aromatic compounds that can be potentially applied as an alternative petroleum-derived feedstock for various bio-based and value-added chemicals. Despite its abundance, the utilization of lignin has been particularly challenging due to its recalcitrant structures (Zhao et al. 2016). Moreover, lignin is a complex amorphous phenylpropanoid polymer that is mainly synthesized from three monomeric precursors (p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units), and those monomers are incorporated in the form of various types of chemical bonds or interunit linkages such as β–O–4, α–O–4/β–5, β–β, 4–O–5, and 5–5 (Munk et al. 2015).
Despite the fact that lignin is highly recalcitrant towards biological degradation, many nature biomass conversion systems, including white rot fungi, bacteria, and wood-feeding termites, are indeed capable of utilizing lignin through the secretion of various lignolytic enzymes (Xie et al. 2014). The microbial degradation of lignin has been well studied in the fungal systems, but it is a real challenge for fungal protein expression and its genetic manipulation. Compared with conventional fungal systems, bacteria have in recent years attracted increased attention due to their unique advantages, such as rapid growth, stable adaptability in a wide range of environmental conditions, and a relatively easygoing genetic manipulation (Bugg et al. 2011; Ma et al. 2016).
Within the past few years, a variety of investigations have been conducted on the role of bacteria in lignin degradation. For example, the bacteria Novosphingobium sp. B-7, Comamonas sp. B-9, Pandoraea sp. B-6, and Cupriavidus basilensis B-8 were successfully isolated from the eroded bamboo slip steeping fluid derived, and those bacteria were identified to have a remarkably higher laccase and peroxidase activity, as well as the ability to aerobically degrade kraft lignin (Chen et al. 2012; Shi et al. 2013a,b; Chai et al. 2014). Among bacteria lignin degraders, Sphingomonas paucimobilis SYK-6 is considered one of the best characterized lignin-degrading bacteria that can utilize a wide variety of lignin-related biaryls and monoaryls as a sole carbon source, and it contains the metabolic pathways for deconstruction of the β-aryl ether, biphenyl, and other components of lignin (Kamimura et al. 2017). Enterobacter lignolyticus SCF1 has the ability to use lignin in both assimilatory and dissimilatory pathways (DeAngelis et al. 2013). The extremophile Bacillus ligniniphilus L1 was able to degrade lignin under the alkali and salty conditions, which was beneficial for the treatment of wastewater containing lignin such as black liquor from the papermaking industry (Zhu et al. 2017). Those reports indicated that those novel lignin-degrading enzymes are mainly to be identified as laccases, lignin peroxidase, glutathione-S-transferase, ring cleaving dioxygenases, monooxygenases, quinone oxidoreductase, esterases, and so on. Compared to lignolytic fungal systems, bacterial systems have been found to be less oxidatively powerful but may provide a plentiful source for illustrating novel accessory enzymes that could act synergistically with the major oxidative enzymes to trigger and uncap various sites (Brown and Chang 2014). Therefore, the exploration of novel bacteria with higher delignification efficiency or a unique degrading strategy may open a new window to learn from a natural mechanism for a better lignin deconstruction process (Sun et al. 2014).
Termites can dissimilate 74% to 99% of the cellulose and 65% to 87% of the hemicelluloses, leaving the modified lignin residues as feces (Li et al. 2017). To date, previous investigations have demonstrated that the fungus-growing termites, as a unique type of wood-feeding termite, have developed a viable mechanism to degrade lignocellulose, particularly with important and complementary assistance from their gut symbionts (Geng et al. 2018). The different types of cellulolytic, hemicellulolytic, and lignolytic symbionts, especially for some symbiotic bacteria interacted with biomass degradation processing in its digestive system, have been successfully isolated and identified (Cibichakravarthy et al. 2017; Tikhe et al. 2017). As a growing research interest, this type of fungus-growing termite may have evolved a unique group of bacterial symbionts to cope with the lignin recalcitrance in their food, which is assumed to be largely dependent on their environment and various food resources. The unique symbiotic relationship between termite hosts and their gut bacteria, which are potentially associated with lignin component depolymerization, remains lacking and unclear in the existing literature. Hence, to better understand a possible function of those involved bacterial symbionts towards lignin component deconstruction, it is essential for the relevant gut bacterial composition profiles, functions, as well as their potential applications for the biorefinery industry to be revealed.
The aim of this investigation was to isolate some novel lignolytic bacterial strains from the gut of a fungus-growing termite species, Microtermes pakistanicus, and further to understand the involved lignin degrading mechanism for these bacterial symbionts assisted with a variety of analyzing tools to characterize their physiochemical properties.
EXPERIMENTAL
Materials
Chemicals and medium
Alkali lignin (AL; catalog #370959), Azure B (AB), Ramazol Brilliant Blue R (RBBR), Methylene blue (MB), and Tetramethylammonium hydroxide (TMAH) were purchased from Sigma-Aldrich (St. Louis, USA). Phenol red (PR) and other chemicals in this study were purchased from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China). The modified mineral salt medium (MSM) used in this study contained (g L-1 deionized water): K2HPO4, 1; KH2PO4, 1; (NH4)2SO4, 2; MgSO4, 0.2; CaCl2, 0.1; FeSO4, 0.05; MnSO4, 0.02; CuSO4, 0.001; ZnSO4, 0.001; and tryptone, 0.05. The medium was sterilized by autoclaving for 15min at 115 °C.
Methods
Isolation and identification of lignin-degrading bacteria from the termite gut
The termites were collected from a decayed tree stump in the tropical rainforest of the Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Xishuangbanna, China. A part of the termite samples was then identified in terms of the sequence of mitochondrial COII gene combined with the soldier sample’s morphology, which has been confirmed as a type of higher termite species, Microtermes pakistanicus.
Simultaneously, 50 worker termites of the M. pakistanicus species were surface sterilized by dipping them in 75% ethanol twice, followed by sterile water, and then they were dried on a piece of clean filter paper for gut system separation. After a dissection process for the termite gut, the internal contents in their gut system were suspended and homogenized in sterile phosphate buffer solution (PBS, pH 7.2, 1 mL). The homogenate samples were made in a serial dilution up to 10-8, and then 100 µL of the final diluted sample was spread on the AL-MSM agar medium. The plates were kept at 30 °C for 3 days and then observed for bacterial growth. The distinct colonies were picked up and purified via repeated spreading (at least three times) on the AL-MSM agar medium, which then were preserved for further study.
The DNA extraction of the bacterial strain, MP-4, was performed using the Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. Amplification of the 16S rRNA gene sequence was performed as described before (Weisburg et al. 1991), and the universal primers 27F and 1492R were used for the polymerase chain reaction. The purified DNA was then packed and sent to Sangong Biotech (Shanghai, China) for sequencing. The sequenced results were compared with the sequences deposited in the GenBank public database from the National Center for Biotechnology Information (NCBI).
Dye decolorization and lignolytic enzyme activity assays
The decolorization of the dyes as an indicator for the production of lignin-degrading enzymes was assessed in solid phase assays. Lignolytic indicator dyes were added to MSM using the methods described by Hooda et al. (2015). The following dyes were tested: PR, AB, RBBR, and MB at concentrations of 100, 50, 50, and 100 mg L-1, respectively. Bacteria were streaked onto dye-containing agar plates and incubated at 30 °C. The plates were observed for growth and decolorization zones after 5 days. Isolates that decolorized any of the dye were screened as positive for the lignolytic enzyme and selected for further lignin degradation studies. Laccase (Lac), lignin peroxidase (LiP) and Mn peroxidase (MnP) assay were performed by the methods as described by Shi (2013 a).
Measurement of bacteria growth and chemical oxygen demand (COD) reduction
Bacterial growth was determined by measuring the culture optical density (OD) at 600 nm with a UV-1200 spectrophotometer (Mapada Instruments, Beijing, China) using an uninoculated medium as a blank. The control and cultured samples were centrifuged at 12,000 rpm for 5 min to collect the supernatant, and then the COD was measured using a 5B-3B type COD analyzer (Lianhua Tech Co., Ltd., Beijing, China). The COD concentration was chosen as the decomposition index to characterize the lignin degradation (Hu et al. 2017). The COD removal ratio was calculated using Eq. 1:
CODremoval (%) = (InitialCOD - FinalCOD) / InitialCOD (1)
Biodegradation of AL
A single colony of MP-4 strain was inoculated into 50 mL of Luria broth (LB) medium and cultivated at 30 °C by shaking at 150 rpm until the OD600 of the inoculums reached approximately 1.0. Then, 2 mL of cells were centrifuged and washed twice with sterile PBS and then were re-suspended with 1 mL AL-MSM medium. The re-suspended cells were inoculated into 200 mL of sterile lignin fermentation medium (AL-MSM, pH 7.0 to 7.2). The fermentation was carried out at 30 °C with a shaking speed of 180 rpm for 7 days.
Analytical Techniques
Field emission scanning electron microscopy (FE-SEM) observation
For the FE-SEM analysis, the control and treated samples were frozen at -80 °C and vacuum-dried into powder. Then, the dried samples were fixed on the double-sided carbon tape, coated with gold-palladium alloy using a JFC-1600 auto fine coater (JEOL Co., Tokyo, Japan), and examined with a JSM-7800F scanning electron microscope (JEOL Co., Tokyo, Japan) operated at an accelerated voltage of 15 kV and the current emission of 84. 5 μA.
Fourier transform infrared (FTIR) characterization
The functional group of characteristics of the control and treated lignin samples were examined by a Nicolet nexus 470 FTIR spectroscope (Thermo Nicolet Corp., Boston, USA). The freeze-dried lignin samples were mixed with KBr at a ratio of 1/100 (w/w) and then milled to a mesh size of 0.1 mm. The powdered samples were made into slices by using a pellet maker (Jingtuo Instrument Science and Technology Ltd., Tianjin, China). The prepared pellets were scanned in the spectra range 4000 to 400 cm−1, and the spectra were recorded using 64 scans per sample at 4 cm−1 resolutions. A pellet prepared with an equivalent quantity of pure KBr powder was used as a background.
Thermogravimetric (TG) analysis
The thermogravimetric/derivative thermogravimetric (TG/DTG) experiments were performed with a TGA 4000 thermogravimetric analyzer (Perkin-Elmer Co., Ltd., Waltham, USA) for the slow pyrolysis of bacteria-treated lignin, as well as a control sample. Approximately 2 mg of the lignin sample were placed into the pan of the TGA microbalance and then were heated from 50 to 800 °C at a heating rate of 20 °C min-1 under low nitrogen of 20 mL min-1. Repeated experiments showed that the TG curves had good reproducibility.
Pyrolysis gas chromatography/mass spectroscopy (GC/MS) analysis for lignin substructure degradation
Pyrolysis-GC/MS was performed as modified by Ke et al. (2013). The pyrolysis processes were performed with a pyrolysis autosampler (model PY-2020Id; Frontier Laboratories, Fukushima, Japan) connected to a QP2010 Ultra GC/MS system (Shimadzu, Kyoto, Japan) equipped with a UA-30M- 0.25F capillary column (30 m in length, 250 µm in inner diameter, and 0.25 µm in film thickness) (Frontier Laboratories, Fukushima, Japan). Approximately 1 mg of each sample (raw lignin and treated lignin) was placed into a quartz tube and mixed with 5 µL of TMAH (25% in methanol, w/w), and then heated up to 610 °C for 30 s. The column used helium (99.999% purity) as the carrier gas (1 mL/min). The temperature program of the GC oven was programmed from 40 °C (hold 1 min) to 280 °C at 8 °C per minute and then held at this temperature for 5 min. The mass spectrometer was operated in electron impact (EI) mode at 70 eV, and the ion-source temperature was 230 °C. The compounds were identified by comparing their GC retention times and mass spectra with the NIST library and reported in the literature. The integration of the peak areas was calculated for each lignin pyrolysis product. The summed areas of the relevant peaks were normalized to 100% and the data were expressed as percentages.
RESULTS AND DISCUSSION
Isolation and Identification of Lignin Degradation Bacteria From the Termite Gut
The lignin-degrading bacteria were isolated in MSM plates with alkali lignin as the sole carbon source. A total of 40 individual bacterial colonies were successfully isolated from the gut of M. pakistanicus and one of the colonies, named as MP-4 (GenBank accession: MF455197.1), presented a noticeable and outstanding degradation ability on the lignin substrate, which was then selected for further investigation regarding its degradation mechanism and characteristics. The 16S RNA sequencing and BLAST analysis showed that the strain MP-4 possessed more than 99% identities with the Pseudocitrobacter anthropi strain C138. In the neighbor-joining phylogenetic tree (Supplementary, Fig. S1), the strain MP-4 fell inside the cluster comprising members of the genus Pseudocitrobacter. Interestingly, as a new genus, the genus Pseudocitrobacter included only two species that were uniquely isolated from the fecal samples of hospitalized patients (Kampfer et al. 2014). However, a similar bacterial strain, MP-4, was the first to be isolated from the gut of a wood-feeding termite and uniquely presented a competitive ability to degrade lignin, which may indicate some distinctive evolution pathway to deal with different environments. As a matter of fact, with this bacterial strain, MP-4, its lignolytic potential is still largely unexplored.
Decolorization Performance with Lignolytic Indicator Dyes and lignolytic enzymes
To confirm that the strain MP-4 was able to produce lignolytic enzymes, the evaluations for a potential decolorization reaction were carried out in an agar plate with one of the four lignin-mimicking dyes, PR, AB, RBBR, or MB as indicators (the structure of dyes given in Fig. S2). The results showed that all four dye substrates were effectively decolorized by the strain MP-4 (Fig. 1). Further, the decolorization reactions with AB, MB, and PR dyes are actually applied to evaluate a potential peroxidase activity (Demidova and Hamblin 2005; Bandounas et al.2011; Yadav and Chandra 2015), while RBBR dye is more reliably used for a laccase promoted activity (Kiiskinen et al. 2004). Therefore, it can be inferred that the strain MP-4 potentially secreted the enzymes of laccase and peroxidase to degrade lignin components. Dye-decolorizing peroxidases (DyPs) in bacteria play a fundamental role in lignin degradation, which are equivalent to lignin peroxidase (LiP) and Mn peroxidase (MnP) in fungi (Bugg et al. 2011). It was speculated that the strain MP-4 might produce laccase and DyPs to support the degradation of lignin. However, it still needs to be further confirmed with a gene sequencing evaluation and an enzyme activity assay.
To better assess the lignolytic potential, three major enzymes (manganese peroxidase, lignin peroxidases and laccases) needed to be quantitatively evaluated. The results of Lac and LiP activity are shown in Fig. S3. MnP activity was not detectable during the 7-day fermentation. Lac and Lip reached maximum activities of 37.66U/L and 86.56 U/L in the lignin cultures, respectively. Compared with previous reported (Ma et al. 2016; Shi et al. 2013a), the strain MP-4 had a lower Lac and LiP activity, and if the enzyme activity is enhanced, optimal fermentation conditions need to be further investigated.
Fig. 1. Agar plate-based decolorization zones around the bacterial colony indicate the degradation process by certain lignolytic enzymes from MP-4 on the tested lignin analogues: a) MB, b) AB, c) PR, and d) RBBR
Bacterial Growth and COD Reduction
To determine the COD reduction and growth of strain MP-4, the bacterial cells were inoculated in a modified MSM medium and cultured for 7 days. The growth curve showed that there was a rapid growth of strain MP-4 at an early stage and until the 4th day; the cells reached a stationary phase between 5 to 7 days, as shown in Fig. 2. Because lignin as the sole organic source contributed to the entire COD load in liquid AL-MSM, the COD reduction could be directly correlated with its lignin degradation. The COD reduction curve indicated that the reduction of COD was not distinct until the 3rd day and then gradually reached its maximum reduction rate at 52.1% after 7 days of growth, which suggested that the degradation of lignin mainly occurred in its growth stationary phase (between 3 to 7 days). Previous reports have showed that the removal rate of COD of strain Cupriavidus basilensis B-8 and Comamonas sp. B-9 were 55.6% and 32%, respectively, with lignin as a single carbon source, after 7 days of fermentation. Similarly, the authors’ results indicated that the MP-4 strain also exhibited a fast growth and efficient degradation rates without any other added carbon sources. These results suggested that the strain MP-4 may demonstrate a potential value in the biorefinery industry.
Fig. 2. Bacterial growth and COD reduction during incubation period in liquid AL-MSM
FE-SEM Analysis
To better understand the deconstruction of lignin by strain MP-4, the morphology of the control and treated lignin samples were investigated by FE-SEM. The micromorphology of the untreated lignin exhibited rigid, compact, and irregular spherical particles with a porous internal structure (Fig. 3a), but the bacteria-treated lignin showed that the structure was completely destructured, and the particle size was reduced from 10 to 100 μm to 2 to 10 μm (Fig. 3b). The changes in the lignin morphology indicated that the lignin had been attacked by strain MP-4.
Fig. 3. FE-SEM images of AL powders. A) Control (untreated) and b) treated lignin for 7 days
FTIR Characterization of Lignin
To understand the chemical functional group changes of alkali lignin during the bacteria depolymerization process, the FTIR spectroscopy analysis was performed to the samples both with the control and the treated lignin after 7 days of incubation. The FTIR spectra of the control and treated samples were apparently different in the typical bands and their intensities (Fig. 4). The assignments of chemical functional groups are listed in Table 1 according to previous references (Jahan et al. 2007; Wang et al. 2009; Yang et al. 2010; Zhao et al. 2014), which can indicate some changes regarding its chemical functional groups.
The absorption band at approximately 3434 to 3234 cm-1 was assigned to the OH groups in the phenolic or aromatic structure. The bands around 2940 to 2938 cm-1 could have corresponded to the asymmetric and symmetrical C–H vibrations of methyl and methylene groups, respectively. The peaks at 1654 cm-1 were assigned to the C=O stretching conjugation with OH groups. In addition, the bands between 1600 to 1400 cm−1 were attributed to the aromatic skeleton vibrations and C–H deformation vibrations, respectively. The presence of the absorption bands at 1270 cm-1 and 1126 cm-1 corresponded to the stretching vibration of syringyl and condensed guaiacyl and guaiacyl rings, respectively, with C=O stretching vibrations that were peaks of a typical guaiacyl-syringyl type lignin. Some characteristic peaks of lignin were found at 1218 cm-1, 1033 cm-1, 854 cm-1, and 816 cm-1, corresponding to C–H or C=O stretching vibrations, aromatic C–H in-plane deformation vibrations, and the C–H out-of-plane stretching vibrations, respectively. Furthermore, these functional groups mainly support the general structure of lignin. Compared with the control samples, the FTIR spectra of the treated samples showed that the relative signal intensity of the absorption (1596 cm-1, 1513 cm-1, 1455 cm-1, and 1425 cm-1) was obviously decreased and the band at 1374 cm-1 disappeared. These results clearly indicated that the strain MP-4 not only caused side chain oxidation and demethylation modifications but also modified and further attacked the aromatic skeletal backbone during the lignin degradation process (Kumar et al. 2015).
In the spectra of the treated samples, the peaks of 854 cm-1 and 816 cm-1 disappeared, and the intensity of the 1126 cm-1 peak was much higher, which indicated that the strain MP-4 deconstructed both the G and S units, and the degradation speed of guaiacyl was faster than syringyl. Moreover, the peak at 1142 cm-1 that originated from the stretch vibration of the ether bond disappeared and the peak at 1084 cm-1 assigned to the C–O deformation at Cβ and aliphatic ether also disappeared. Notably, the disappearance of some bond linkages, such as β–O–4 and Cβ, suggested that the strain MP-4 presented a remarkable capability to deconstruct the lignin components, simply because the linkages of β–O–4 are the most important linkages between monomer units of lignin that account for approximately 50% to 70% of linkages.
Fig. 4. FTIR spectra of the control and treated lignin substrates after 7 days
Table 1. Assignment of FTIR Spectra from Control and Degraded Lignin Samples as Given in Fig. 4
Thermal Analysis
In general, the thermal properties of lignin are related to its highly aromatic backbone and its thermal decomposition that occurs with bond cleaved reactions at different temperatures depending on bond energies (Ke and Chen 2013). The TG and DTG curves at heating rates of 20 °C /min for the treated lignin by stain MP-4 and the control group were obtained and depicted in Fig. 5. Usually, the first stage of weight loss (2% to 6%) at 100 to 180 °C was assigned to the residual moisture evaporation. The treated lignin samples started to decompose from 180 °C and ended at 600 °C, while the control samples started to decompose at 200 °C and showed gradual mass loss until 800 °C. Meanwhile, the treated samples had a higher char residue (54.7%) at the final temperature than that of the control (29.3%), which suggested that the lignin structure was already modified during the 7 days of reaction processing by the strain MP-4.
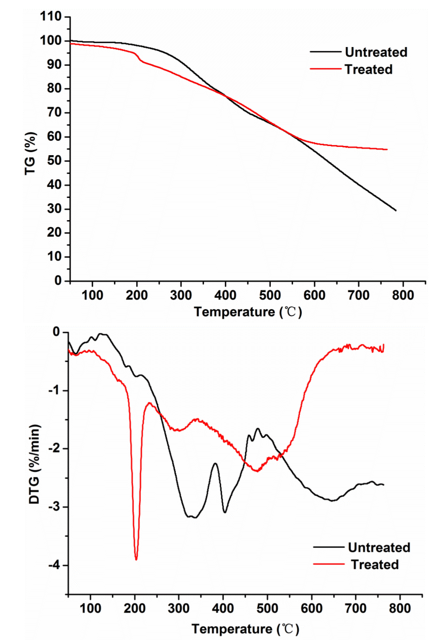
Fig. 5. TG and DTG reaction curves on the lignin substrates after treatment for pyrolysis at a heating rate of 20 °C/min
Great differences were observed between the treated samples and the controls in the DTG curves. The three major mass loss peaks of the treated lignin appeared at 201 °C, 300 °C, and 478 °C but the loss peaks of the control groups appeared at 330 °C, 400 °C, and 640 °C, respectively. The thermal decomposition rate of those treated lignin samples was higher than that of the control samples in the early decomposition stage (200 to 350 °C), which was primarily attributed to the cleavage of lignin side chains and weaker bonds, such as β–γ carbon bonds in the side chain, –OH linked to β or γ carbon, and the C–O bond in the β–O–4′ structure (Lin et al. 2015). In the stage of decomposition at 350 to 400 °C, the increased decomposition rate was due to the oxidation of the lignin side chain (i.e., carbonylation/carboxylation of the aliphatic hydroxyl group and side chain dehydrogenation) (Shen et al. 2016). Afterwards, when the temperature reached above 400 °C, the mass loss of lignin was attributed to the cleavage of C–C linkages (mainly 5–5′ and β–β’) within the lignin skeleton and the demethoxylation reaction truly occurred from 400 °C to 500 °C (Sahoo et al. 2011; Liu et al. 2016). Clearly, the control and treated lignin samples displayed markedly different thermal decomposition behaviours, suggesting that the strain MP-4 would remarkably modify the lignin structures.
Py-GC/MS Analysis
To investigate other structural changes of the lignin substrates induced by the degradation process from the strain MP-4, the Py-GC/MS analysis was further performed with the treated lignin after 7 days of incubation. The pyrolysis of the treated lignin and control samples was conducted at 610 °C and yielded a total of 78 derivatives, including phenol, 2-methylphenol, 3-methylphenol, 4-methylphenol, 4-methoxyphenol, 3-dimethylphenol, 2,5-dimethylphenol, 1,2-dimethoxybenzene, 2,4-dimethylphenol, 2,3,5-trimethylphenol, 2,3-dimethoxytoluene, 2,3-dihydroxytoluene, methyleugenol, 2′,4′-dihydroxy-3′-methylpropio-phenone, etc. (Fig. 6). The predominant derivative products were phenolic compounds, which implied that the aryl ether linkages and C–C bonds were possibly cleaved. Table 2 summarizes the major pyrolysis products, where their corresponding peak areas were also listed. These pyrolysates were composed of H-, G-, and S-type lignin derivatives, which was consistent with previous reports (Ke et al. 2011; Chu et al. 2013; Zhang et al. 2014; Yan et al. 2016).
As shown in Fig. 6, the pyrolysate profiles of lignin treated with or without bacteria processing produced quantitatively similar pyrolysis products, but clear visible differences in the amounts of the pyrolysates were found. For example, 4-methyl-phenol (peak 8) and 4-methoxy-phenol (peak 9) showed the highest relative abundances in the pyrolysates of the untreated lignin samples, but they disappeared after the bacterial treatment. Likewise, some lignin-derived compounds, such as peak 2, 11, 16, 20, 22, 23, 25, 27, 29, 35, 36, 39, 44, 45, 47, 53, 55, 60, 61, 63, 67 through 70, 72, and 75 through 77, completely disappeared. In addition, a variety of new derivative compounds (peaks 7, 10, 13, 15, 21, 26, 28, 30, 32, 34, 40, 52, 54, 56, and 58) were generated in the program of the degraded lignin compared with the control samples. This indicated that the lignin degradation or modification by strain MP-4 occurred in the chemical structure and/or interunit level. The disappearance of pyrolysates 67 to 69 and 76, which were potentially derived from the lignolytic biphenyl structure, implied a plausible cleavage of C–C/C–O within the lignin 5–5′ substructure. The relative contents of 2-methoxy-4-propyl-phenol (peak 33) decreased in those treated samples when compared with its raw lignin samples, which may demonstrate an acylating process on the side chains of the lignin polymers via decarboxylation degradation (Del Rio et al. 2007). Moreover, the relative intensity of pyrolysate 19 and 78 has been also changed. Among of them, pyrolysate 19 was considered to be derived from the β–O–4′ substructure, while pyrolysate 78 could have possibly been generated from the lignin β–β and β–5′ substructures. These results have clearly provided evidence for lignin substructure modifications at sites of β–O–4′, β–1′, and β–5′ after bacterial treatment (Kuroda et al. 2002; Zeng et al. 2013). The disappearance of pyrolysate 8 (H unit) in the treated lignin was the most intense peak in the untreated control groups and in contrast, the pyrolysate 9 (G unit), 16 (H unit), 20 (H unit), 25 (S unit), and 29 (G unit) were not observed in the treated samples, indicating that the degradation of lignin was at the sub-structural and/or interunit level by the strain MP-4.
As for the pyrolysates from those untreated lignin samples, the total peak areas of H, G, and S lignin derivatives were recorded at 28.6%, 31.28%, and 18.73%, respectively. However, after bacterial processing, the total peak areas of H, G, and S lignin derivatives were recorded at 28.73%, 31.9%, and 8.08%, respectively. Compared with those untreated samples, the total yields of the G-type and H-type derivatives slightly increased. However, the S-type derivative yields from the treated samples were remarkably reduced, and the relative peak areas decreased 56.86% after the bacterial treatment. Meanwhile, the S/G ratio decreased from 0.59 to 0.25. Therefore, these results showed that the S-type lignin unit was much easier to be attacked than that of the G-type lignin unit due to its lower redox potential, and a linear arrangement of the S units by β–O–4′ linkages that were more attackable than the G units (Yan et al.2016).
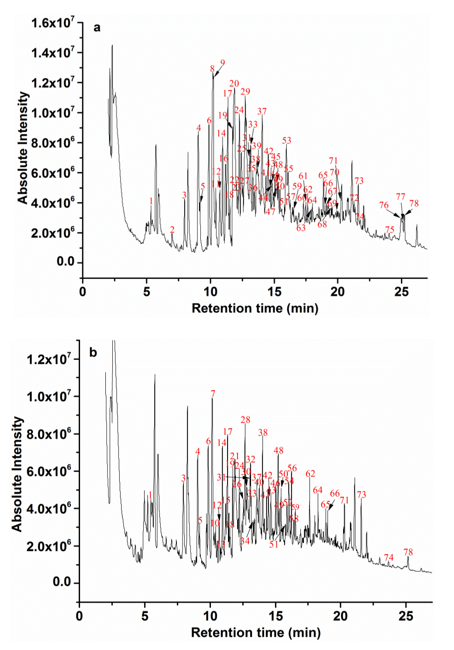
Fig. 6. Py–GC/MS programs of lignin at 610 °C: a) untreated and b) treated samples
Table 2. Pyrolysis Product Analysis of Lignin Derived From the Untreated and Bacterial Pretreated Samples
S- syringyl-type lignin derivatives, G- guaiacyl-type lignin derivatives, and H- p-hydroxy phenylpropane -type lignin derivatives; ND- not detected
Proposed Degradation Mechanism of Lignin
As discussed above, the lignin depolymerization mechanism was proposed as follows (Fig. 7): strain MP-4 initially broke down the lignin structure through cleaving β–O–4 linkages as well as C-C bonds to form lower molecular weight aromatic compounds; then, these aromatic compounds were utilized as carbon and energy sources by Pseudocitrobacter anthropi MP-4 through β-ketoadipate (β-KAP) pathway (Shi et al. 2013a). The similar depolymerization mechanisms on lignin degradation were also reported in other bacterial species, including Sphingomonas paucimobilis SYK-6 (Kamimura et al. 2017), Enterobacter lignolyticus SCF1 (DeAngelis et al. 2013), and Cupriavidus basilensis B-8 (Shi et al. 2013a). These bacteria would commonly break down the lignin polymer substrates into a small aromatic molecule that could be then utilized by bacterial cells for a final decomposition.
With respect to this mechanism, it can be supported by the FTIR or Py-GC/MS analyses on those lignin substrates before and after the strain MP-4 interactions. In the authors’ case, the FTIR spectra exhibited some noticeable differences in their absorption peaks between the control and treated lignin substrates with the bacterial strain MP-4. The peaks at 816, 854, 1031, 1084, and 1142 cm-1 in the treated groups completely disappeared, indicating that the β–O–4′ substructure was destroyed by strain MP-4. Further analysis also indicated that the intensity band at 2938 and 1142 cm-1 decreased, suggesting that a demethoxylation/demethylation reaction probably occurred after the bacterial treatment. The decreased intensities at 1596 and 1425 cm-1 further indicated that the aromatic skeletal carbon of lignin was destroyed by strain MP-4. In addition, the increased intensity of the peak at approximately 1654 cm-1 was assigned to the C=O stretching conjugation with carboxylic acid and ketone groups, which could be due to a side chain oxidation or ring opening and formation of an additional carbonyl group (Bule et al. 2013).
Fig. 7. A proposed degradation scheme made by strain MP-4 on lignin substrates
For Py-GC/MS analysis, the pyrolysates (peaks 67, 68, 69, and 76) in the pyrolysis program for those treated lignin substrates also completely disappeared, which implied that the carbon-carbon bonds were possibly cleaved. In summary, the mechanisms on lignin substrate degradation by bacterial strain MP-4 can be uniquely combined with a variety of reactions, including the demethoxylation/demethylation, the cleavage of β–O–4′ aryl ether, C–C bonds, as well as the side chain oxidation.
CONCLUSIONS
- In this study, the novel lignin-degrading bacterial strain Pseudocitrobacter anthropi MP-4 was successfully isolated from the gut of the wood-feeding termite species, M. pakistanicus. Pseudocitrobacter anthropi MP-4 exhibited a fast growth and efficient degradation rates in the lignin medium, and had the ability to produce laccase (Lac) and lignin peroxidise (Lip) enzymes.
- The characteristics in lignin degradation by this novel bacterial strain were further identified by various analyses and observations via SEM, FTIR, TGA, and Py-TMAH-GC/MS methods. The results showed that the structure and thermal stability of lignin had a notable change via the degradation process from the bacterial strain MP-4.
- A possible depolymerization mechanism of lignin was proposed as follows: the strain MP-4 initially broke down the lignin structure through cleaving β–O–4 linkages as well as C-C bonds to form small molecular aromatic compounds; then, these aromatic compounds were mineralized by Pseudocitrobacter anthropi MP-4.
ACKNOWLEDGMENTS
This research was funded by the National Natural Science Foundation of China (No. 31772529), the Research Innovation Program for College Graduates of Jiangsu Province (No. KYLX16_0910), a project funded by the Priority Program Development of Jiangsu Higher Education Institutions.
REFERENCES CITED
Bandounas, L., Wierckx, N. J. P., De Winde, J. H., and Ruijssenaars, H. J. (2011). “Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential,” BMC Biotechnol. 11, 94-104. DOI: 10.1186/1472-6750-11-94
Brown, M. E., and Chang, M. C. Y. (2014). “Exploring bacterial lignin degradation,” Curr. Opin. Chem. Biol. 19, 1-7. DOI: 10.1016/j.cbpa.2013.11.015
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., and Singh, R. (2011). “The emerging role for bacteria in lignin degradation and bio-product formation,” Curr. Opin. Biotech. 22(3), 394-400. DOI: 10.1016/j.copbio.2010.10.009
Bule, M. V., Gao, A. H., Hiscox, B., and Chen, S. L. (2013). “Structural modification of lignin and characterization of pretreated wheat straw by ozonation,” J. Agr. Food Chem. 61(16), 3916-3925. DOI: 10.1021/jf4001988
Chai, L. Y., Chen, Y. H., Tang, C. J., Yang, Z. H., Zheng, Y., and Shi, Y. (2014). “Depolymerization and decolorization of kraft lignin by bacterium Comamonas sp. B-9,” Appl. Microbiol. Biot. 98(4), 1907-1912. DOI: 10.1007/s00253-013-5166-5
Chen, Y. H., Chai, L. Y., Tang, C. J., Yang, Z. H., Zheng, Y., Shi, Y., and Zhang, H. (2012). “Kraft lignin biodegradation by Novosphingobium sp. B-7 and analysis of the degradation process,” Bioresource Technol. 123, 682-685. DOI: 10.1016/j.biortech.2012.07.028
Chu, S., Subrahmanyam, A. V., and Huber, G. W. (2013). “The pyrolysis chemistry of a beta-O-4 type oligomeric lignin model compound,” Green Chem. 15(1), 125-136. DOI: 10.1039/C2GC36332A
Cibichakravarthy, B., Abinaya, S., and Prabagaran, S. R. (2017). “Syntrophic association of termite gut bacterial symbionts with bifunctional characteristics of cellulose degrading and polyhydroxyalkanoate producing bacteria,” Int. J. Biol. Macromol. 103, 613-620. DOI: 10.1016/j.ijbiomac.2017.05.100
DeAngelis, K. M., Sharma, D., Varney, R., Simmons, B., Isern, N. G., Markilllie, L. M., Nicora, C., Norbeck, A. D., Taylor, R. C., Aldrich, J. T., et al. (2013). “Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1,” Front. Microbiol. 4, Article 280. DOI: 10.3389/fmicb.2013.00280
Del Rio, J. C., Gutierrez, A., Rodriguez, I. M., Ibarra, D., and Martinez, A. T. (2007). “Composition of non-woody plant lignins and cinnamic acids by Py-GC/MS, Py/TMAH and FTIR,” J. Anal. Appl. Pyrol. 79(1-2), 39-46. DOI: 10.1016/j.jaap.2006.09.003
Demidova, T. N., and Hamblin, M. R. (2005). “Photodynamic inactivation of Bacillus spores, mediated by phenothiazinium dyes,” Appl. Environ. Microb. 71(11), 6918-6925. DOI: 10.1128/AEM.71.11.6918-6925.2005
Geng, A., Cheng, Y. B., Wang, Y. L., Zhu, D. C., Le, Y. L., Wu, J., Xie, R. R., Yuan, J. S., and Sun, J. Z. (2018). “Transcriptome analysis of the digestive system of a wood-feeding termite (Coptotermes formosanus) revealed a unique mechanism for effective biomass degradation,” Biotechnol. Biofuels 11, 24-37. DOI: 10.1186/s13068-018-1015-1
Hooda, R., Bhardwaj, N. K., and Singh, P. (2015). “Screening and identification of ligninolytic bacteria for the treatment of pulp and paper mill effluent,” Water Air Soil Poll. 226, 305-315. DOI: 10.1007/s11270-015-2535-y
Hu, L. H., Liu, X. R., Wang, Q. X., and Zhou, Y. L. (2017). “Highly efficient degradation of high-loaded phenol over Ru-Cu/Al2O3 catalyst at mild conditions,” RSC Adv. 7(35), 21507-21517. DOI: 10.1039/C7RA00545H
Jahan, M. S., Chowdhury, D. A. N., Islam, M. K., and Moeiz, S. M. I. (2007). “Characterization of lignin isolated from some nonwood available in Bangladesh,” Bioresource Technol.98(2), 465-469. DOI: 10.1016/j.biortech.2006.01.005
Kamimura, N., Takahashi, K., Mori, K., Araki, T., Fujita, M., Higuchi, Y., and Masai, E. (2017). “Bacterial catabolism of lignin‐derived aromatics: New findings in a recent decade: Update on bacterial lignin catabolism,” Env. Microbiol. Rep. 9(6), 679-705. DOI: 10.1111/1758-2229.12597
Kampfer, P., Glaeser, S. P., Raza, M. W., Abbasi, S. A., and Perry, J. D. (2014). “Pseudocitrobacter gen. nov., a novel genus of the Enterobacteriaceae with two new species Pseudocitrobacter faecalis sp nov., and Pseudocitrobacter anthropi sp nov, isolated from fecal samples from hospitalized patients in Pakistan,” Syst. Appl. Microbiol. 37(1), 17-22. DOI: 10.1016/j.syapm.2013.08.003
Ke, J., and Chen, S. L. (2013). “Thermal decomposition of lignin structural modification in termite digested softwood (II),” Fuel 104, 781-787. DOI: 10.1016/j.fuel.2012.06.066
Ke, J., Laskar, D. D., and Chen, S. L. (2011). “Biodegradation of hardwood lignocellulosics by the western poplar clearwing borer, Paranthrene robiniae (Hy. Edwards),” Biomacromolecules 12(5), 1610-1620. DOI: 10.1021/bm2000132
Ke, J., Laskar, D. D., and Chen, S. L. (2013). “Tetramethylammonium hydroxide (TMAH) thermochemolysis for probing in situ softwood lignin modification in each gut segment of the termite,” J. Agr. Food Chem. 61(6), 1299-1308. DOI: 10.1021/jf3048548
Kiiskinen, L. L., Ratto, M., and Kruus, K. (2004). “Screening for novel laccase-producing microbes,” J. Appl. Microbiol. 97(3), 640-646. DOI: 10.1111/j.1365-2672.2004.02348.x
Kumar, M., Singh, J., Singh, M. K., Singhal, A., and Thakur, I. S. (2015). “Investigating the degradation process of kraft lignin by beta-proteobacterium, Pandoraea sp. ISTKB,” Environ. Sci. Pollut. R. 22(20), 15690-15702. DOI: 10.1007/s11356-015-4771-5
Kuroda, K., Nakagawa-Izumi, A., and Dimmel, D. R. (2002). “Pyrolysis of lignin in the presence of tetramethylammonium hydroxide (TMAH): Products stemming from beta-5 substructures,” J. Agr. Food Chem. 50(12), 3396-3400. DOI: 10.1021/jf011563c
Li, H. J., Yelle, D. J., Li, C., Yang, M. Y., Ke, J., Zhang, R. J., Liu, Y., Zhu, N., Liang, S. Y., Mo, X. C., et al. (2017). “Lignocellulose pretreatment in a fungus-cultivating termite,” P. Natl. Acad. Sci. USA. 114(18), 4709-4714. DOI: 10.1073/pnas.1618360114
Lin, X. N., Sui, S. J., Tan, S., Pittman, C. U., Sun, J. P., and Zhang, Z. J. (2015). “Fast pyrolysis of four lignins from different isolation processes using Py-GC/MS,” Energies 8(6), 5107-5121. DOI: 10.3390/en8065107
Liu, C., Hu, J., Zhang, H. Y., and Xiao, R. (2016). “Thermal conversion of lignin to phenols: Relevance between chemical structure and pyrolysis behaviors,” Fuel 182, 864-870. DOI: 10.1016/j.fuel.2016.05.104
Ma, J. S., Zhang, K. K., Liao, H. D., Hector, S. B., Shi, X. W., Li, J. L., Liu, B., Xu, T., Tong, C. Y., Liu, X. M., et al. (2016). “Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd-1,” Biotechnol. Biofuels 9, 25-39. DOI: 10.1186/s13068-016-0439-8
Munk, L., Sitarz, A. K., Kalyani, D. C., Mikkelsen, J. D., and Meyer, A. S. (2015). “Can laccases catalyze bond cleavage in lignin?,” Biotechnol. Adv. 33(1), 13-24. DOI: 10.1016/j.biotechadv.2014.12.008
Sahoo, S., Seydibeyoglu, M. O., Mohanty, A. K., and Misra, M. (2011). “Characterization of industrial lignins for their utilization in future value added applications,” Biomass Bioenerg.35(10), 4230-4237. DOI: 10.1016/j.biombioe.2011.07.009
Shen, X. J., Wang, B., Huang, P. L., Wen, J. L., and Sun, R. C. (2016). “Understanding the structural changes and depolymerization of Eucalyptus lignin under mild conditions in aqueous AlCl3,” RSC. Adv. 6(51), 45315-45325. DOI: 10.1039/C6RA08945C
Shi, Y., Chai, L. Y., Tang, C. J., Yang, Z. H., Zhang, H., Chen, R. H., Chen, Y. H., and Zheng, Y. (2013a). “Characterization and genomic analysis of kraft lignin biodegradation by the beta-proteobacterium Cupriavidus basilensis B-8,” Biotechnol. Biofuels 6, 1-14. DOI: 10.1186/1754-6834-6-1
Shi, Y., Chai, L. Y., Tang, C. J., Yang, Z. H., Zheng, Y., Chen, Y. H., and Jing, Q. X. (2013b). “Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips,” Bioproc. Biosyst. Eng. 36(12), 1957-1965. DOI: 10.1007/s00449-013-0972-9
Sun, J. Z., Ding, S. Y., and Peterson, J. D. (2014). Biological Conversion of Biomass for Fuels and Chemicals–Explorations from Natural Utilization Systems, RSC Publishing, Cambridge, UK. DOI: 10.1039/9781849734738
Tikhe, C. V., Sethi, A., Delatte, J., and Husseneder, C. (2017). “Isolation and assessment of gut bacteria from the Formosan subterranean termite, Coptotermes formosanus (Isoptera: Rhinotermitidae), for paratransgenesis research and application,” Insect Sci. 24(1), 93-102. DOI: 10.1111/1744-7917.12282
Wang, S. R., Wang, K. G., Liu, Q., Gu, Y. L., Luo, Z. Y., Cen, K. F., and Fransson, T. (2009). “Comparison of the pyrolysis behavior of lignins from different tree species,” Biotechnol. Adv. 27(5), 562-567. DOI: 10.1016/j.biotechadv.2009.04.010
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). “16S ribosomal DNA amplification for phylogenetic study,” J. Bacteriol. 173(2), 697-703. DOI: 10.1128/jb.173.2.697-703.1991
Xie, S. X., Syrenne, R., Sun, S., and Yuan, J. S. (2014). “Exploration of natural biomass utilization systems (NBUS) for advanced biofuel – from systems biology to synthetic design,” Curr. Opin. Biotech. 27, 195-203. DOI: 10.1016/j.copbio.2014.02.007
Yadav, S., and Chandra, R. (2015). “Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry,” J. Environ. Sci. 33, 229-238. DOI: 10.1016/j.jes.2015.01.018
Yan, K. L., Liu, F., Chen, Q., Ke, M., Huang, X., Hu, W. Y., Zhou, B., Zhang, X. Y., and Yu, H. B. (2016). “Pyrolysis characteristics and kinetics of lignin derived from enzymatic hydrolysis residue of bamboo pretreated with white-rot fungus,” Biotechnol. Biofuels 9, 76-86. DOI: 10.1186/s13068-016-0489-y
Yang, X. W., Ma, F. Y., Zeng, Y. L., Yu, H. B., Xu, C. Y., and Zhang, X. Y. (2010). “Structure alteration of lignin in corn stover degraded by white-rot fungus Irpex lacteus CD2,” Int. Biodeter. Biodegr. 64(2), 119-123. DOI: 10.1016/j.ibiod.2009.12.001
Zeng, J., Singh, D., Laskar, D. D., and Chen, S. (2013). “Degradation of native wheat straw lignin by Streptomyces viridosporus T7A,” Int. J. Environ. Sci. Te. 10(1), 165-174. DOI: 10.1007/s13762-012-0085-z
Zhang, M., Resende, F. L. P., and Moutsoglou, A. (2014). “Catalytic fast pyrolysis of aspen lignin via Py-GC/MS,” Fuel 116, 358-369. DOI: 10.1016/j.fuel.2013.07.128
Zhao, C., Xie, S. X., Pu, Y. Q., Zhang, R., Huang, F., Ragauskas, A. J., and Yuan, J. S. (2016). “Synergistic enzymatic and microbial lignin conversion,” Green Chem. 18(5), 1306-1312. DOI: 10.1039/C5GC01955A
Zhao, J., Wang, X. W., Hu, J., Liu, Q., Shen, D. K., and Xiao, R. (2014). “Thermal degradation of softwood lignin and hardwood lignin by TG-FTIR and Py-GC/MS,” Polym. Degrad. Stabil. 108, 133-138. DOI: 10.1016/j.polymdegradstab.2014.06.006
Zhu, D. C., Zhang, P. P., Xie, C. X., Zhang, W. M., Sun, J. Z., Qian, W. J., and Yang, B. (2017). “Biodegradation of alkaline lignin by Bacillus ligniniphilus L1,” Biotechnol. Biofuels 10, 44-57. DOI: 10.1186/s13068-017-0735-y
Article submitted: October 1, 2018; Peer review completed: December 15, 2018; Revised version received and accepted: January 17, 2019; Published: January 23, 2019.
DOI: 10.15376/biores.14.1.1992-2012
APPENDIX
Fig. S1. Neighbour-joining tree based on 16S rRNA gene sequences showing the phylogenetic position of strain MP-4 and related species
Fig. S2. The chemical structure of Methylene blue (MB), Azure B (AB), Phenol red (PR), and Ramazol Brilliant Blue R (RBBR)
Fig. S3. The time course of laccase (Lac) and lignin peroxidise (Lip) activities in the culture of lignin
