Abstract
This study aims to complement previous studies on down-regulation of sucrose synthase and cinnamate 4-hydroxylase in hybrid aspen (Bjurhager et al. 2010; Gerber et al. 2014) with a quantitative anatomical comparison (Sandquist et al. 2015). The main focus was placed on evaluating quantitative modifications to fiber morphology and lignin composition. This was achieved by combining results from light-, electron-, and UV-microscopy together with histochemical staining. Overall there was good agreement between the morphological results in this study compared to the previously published chemical and physical reports, particularly on cinnamate 4-hydroxylase down-regulated aspen. The previously reported statistical correlations between fiber lumen area and plant diameter, and fiber length and tree height (Sandquist et al. 2015) have been further refined. Knock-down of genes coding for biologically important proteins such as sucrose synthase and cinnamate 4-hydroxylase had broad spanning morphological impacts. Down-regulated sucrose synthase genotype showed significantly reduced fiber development, possibly with altered S/G ratio. Down-regulated cinnamate 4-hydroxylase genotype showed an overall reduction of lignin in fibers as indicated from both chemical staining and UV-microscopy measurements. Down-regulated pectate lyase genotype showed no overall significant effects on fiber morphology, ultrastructural, nor lignin staining.
Download PDF
Full Article
Anatomical and Immunocoverage Observations on SuSy, C4H, and Pectate Lyase Family Protein Down-regulated Aspens Genotypes
David Sandquist, a Lennart Norell, b Keiji Takabe, c Arata Yoshinaga, c and Geoffrey Daniel d*
This study aims to complement previous studies on down-regulation of sucrose synthase and cinnamate 4-hydroxylase in hybrid aspen (Bjurhager et al. 2010; Gerber et al. 2014) with a quantitative anatomical comparison (Sandquist et al. 2015). The main focus was placed on evaluating quantitative modifications to fiber morphology and lignin composition. This was achieved by combining results from light-, electron-, and UV-microscopy together with histochemical staining. Overall there was good agreement between the morphological results in this study compared to the previously published chemical and physical reports, particularly on cinnamate 4-hydroxylase down-regulated aspen. The previously reported statistical correlations between fiber lumen area and plant diameter, and fiber length and tree height (Sandquist et al. 2015) have been further refined. Knock-down of genes coding for biologically important proteins such as sucrose synthase and cinnamate 4-hydroxylase had broad spanning morphological impacts. Down-regulated sucrose synthase genotype showed significantly reduced fiber development, possibly with altered S/G ratio. Down-regulated cinnamate 4-hydroxylase genotype showed an overall reduction of lignin in fibers as indicated from both chemical staining and UV-microscopy measurements. Down-regulated pectate lyase genotype showed no overall significant effects on fiber morphology, ultrastructural, nor lignin staining.
Keywords: Wood genotyping; Immunocoverage; Wood ultrastructure; Wood Chemistry; Cinnamate 4-hydroxylase; Sucrose synthase; UV-microscopy; Quantitative microscopy
Contact information: a: University of Hamburg, Leuschnerstr. 91c, 21031 Hamburg, Germany; b: Applied Statistics and Mathematics, Department of Economics, Swedish University of Agricultural Sciences, P.O. Box 7013, SE-750 07 Uppsala, Sweden; c: Tree Cell Biology, Division of Forest and Biomaterials Science, Kyoto University, Oiwake-cho, Kitashirakawa, Kyoto 606-8502, Japan; d: Wood Science, Department of Forest Products, Swedish University of Agricultural Sciences, P.O. Box 7008, SE-750 07 Uppsala, Sweden;
* Corresponding author: geoffrey.daniel@slu.se
INTRODUCTION
The increasing world demand for plant lignocellulosic biomass has accelerated in recent years as the competition for conventional use of wood material for pulp and paper and construction has been further extended as a resource for bioenergy and more recently biofuels (Vanholme et al. 2008). This competition for lignocellulose sees no sign of plateauing out but rather is likely to accelerate as resources begin to dwindle and pressure from accompanying environmental concerns increase. The ability to “domesticate” trees through genetic engineering has great potential as a means for producing new clones for producing greater or modified biomass in a shorter time via enhanced growth (Hu et al.1999; Pilate et al. 2002). Most of the early research on genetic engineering of trees has been conducted using poplar as a model system due to its rapid growth rate and the fact that its genome has been fully sequenced (Tuskan et al. 2003), simplifying the production of transgenic plants (Hu et al. 1999; Boerjan et al. 2003; Li et al. 2003).
Wood is composed primarily of cellulose, lignin, and hemicelluloses, as well as other minor components (Sjöström 1993). Of these, cellulose provides load-bearing in trees and products, while lignin gives wood its hydrophobic character, natural resistance to compressive and tension forces, as well as protection against decay (Fengel and Wegener 1989). Lignin also provides rigidity to wood and at the macromolecular level is cross-linked to the hemicelluloses (Sjöström 1993).
Lignin is a recalcitrant component in wood that needs to be removed during chemical pulping, a process that is both energy-consuming (requiring heat and chemicals) and environmentally negative (Hu et al. 1999; Chiang 2002; Li et al. 2003; Baucher et al. 2003; Vanholme et al. 2008). The major industrial potential of the modification/reduction of lignin in trees in both content and composition was thus realized quite early on and much research has since been orientated to understanding the biosynthetic pathways of lignin metabolism in trees with this purpose in mind (Boerjan et al. 2003; Baucher et al. 2003; Vanholme et al. 2008). One of the major early findings was demonstration in transgenic poplar that repression of lignin biosynthesis also promotes cellulose accumulation and growth (Hu et al. 1999; Li et al. 2003).
Despite the intense research on lignin biosynthesis in poplar species, few studies appear however to have concentrated on the morphological effects on wood anatomy, and spatial changes in lignin in relation to ultrastructure and physical properties caused by modifications in lignin biosynthesis (Leple et al. 2007; Horvath et al. 2010).
The aim of this study was to investigate changes induced in the wood anatomical (tissue) structure, lignin micro-distribution, and ultrastructure of three transgenic aspen trees (Populus tremula x tremuloides). Each tree has one knocked down gene each, abbreviated as KR112 (sucrose synthase, Sjödin et al. 2009), KR139 (cinnamate 4-hydroxylase, Gerber et al. 2014) and K213 (pectate lyase coding genes, Sandquist et al. 2015) in comparison with their reference wild type control (T89).
The study complements and extends previously published observations on the three genotypes (Table 1), with detailed anatomical structural observations. This has been achieved using quantitative image analysis of transverse sections (Sandquist et al. 2015) and macerated fibers, UV-microscopy and histochemical staining of syringyl- and guaiacyl lignin micro-distributions, and ultrastructural observations on changes in fiber wall structure as determined using TEM.
EXPERIMENTAL
Materials
Genetic transformation
Antisense constructs were generated by cloning a partial cDNA (CloneID, popgenie.org, Sjödin et al.2009) as shown in Table 1 in a gateway compatible anti-sense binary vector, pK7GWIWG2(I) (Karimi et al. 2002). The construct was then transformed into hybrid aspen (clone T89) (Populus tremula x tremuloides) as described previously (Nilsson et al. 1992). Kanamycin-resistant lines were clonally propagated in vitro and planted in a greenhouse.
Table 1. Aspen Genotype Constructs Used in the Study
Plant Material
Eighteen hybrid transgenic aspen (Populus tremula x tremuloides) were grown in a greenhouse with an 18-h photo-period, a temperature regime of 22/17 ºC (day/night) and relative humidity (RH) of at least 70%. Natural daylight was supplemented with light from HQI-TS 400W/DH metal halogen lamps (Osram, Munich, Germany). Plants were watered daily and fertilized once per week with a 1:100 dilution of SUPERBA S (Hydro Supra AB, Landskrona, Sweden). For each of the three genotypes there were two independent lines, with three replicates. The genotypes were accompanied with six non-transformed reference T89 trees. Trees were planted in February 2010 and harvested at 3 months of age. Plants were measured for height and diameter weekly.
Methods
Sampling, embedding, and sectioning
From each tree at 20 cm above soil height, a 1 cm high section of stem was collected, cut into ~2 mm sectors, and fixed with 3% v/v glutaraldehyde containing 2% v/v paraformaldehyde in cacodylate buffer (0.1 M, pH 7.2) for 9 h at room temperature. Three 1×1 mm samples were collected from the primary fixed sector for each genotype and post-fixed with osmium tetroxide (2% w/v in cacodylate buffer) for 2 h at room temperature. After fixation the samples were washed (2×20 min) with distilled water and dried with an ethanol series consisting of 20 min steps (20, 40, 60, 70, 80, 90, 95, and 99.5%).
For each tree, 1×1 mm primary fixed, samples were cut from the fixed sectors and embedded in Technovit 8100 (Kulzer, Wehrheim, Germany), LR White (London Resin, Basingstoke, UK), and Spurr’s resin (Spurr 1969; Agar Scientific, Stansted, UK) according to the manufacturer instructions. The three post-fixed samples were embedded similarly in LR White. For all resins, infiltration solution concentrations were gradually increased over one week.
From the embedded Technovit 8100 samples, 2 µm transverse sections were collected on a Microm microtome (HM 350, Microm, Germany). From the Spurr’s resin embedded samples 1 µm transverse sections were collected on a Reichert-Jung Ultracut E ultra-microtome. With the LR White embedded samples, silver and gold ultrathin transverse sections were collected using a Reichert-Jung Ultracut E ultra-microtome and collected on 100 and 200 mesh nickel grids for TEM.
Wood maceration
Three sectors of wood, approx. 3 mm by 1 cm for each tree were delignified and macerated by treatment in a 1:1 mixture of 100% acetic acid and H2O2 at 60 ºC for 18 h to allow fiber length and width measurements (Wise and Murphy 1946).
Chemical staining
Toluidine blue staining was used as an aid for morphological characterization and measurements on transverse Technovit embedded samples (Chaffey 2002). All samples were control stained to verify the absence of tension wood by dual staining with Safranin and Astra Blue (Chaffey 2002). Phloroglucinol and Mäule staining were carried out on fresh 10 µm wood sections and non-resin embedded sections, according to procedures given by Chaffey (2002).
UV-microscopy
Transverse 1 µm Spurr’s resin sections were mounted using glycerin on quartz slides and covered with quartz coverslips. UV-absorption was measured at 280 nm using a Carl Zeiss UMSP 80 microscopic spectrophotometer, mainly reflecting guaiacyl absorption (Yoshinaga and Wada 2007). UV-absorption was measured across double fiber-fiber secondary cell walls using ImageJ (Abramoff et al. 2004). The mean absorption was taken as the maximum for 50 fiber-fiber cell walls per sample.
Transmission electron microscopy
Ultrathin osmicated sections were post-stained with 2% w/v potassium permanganate solution containing sodium citrate. Sections were observed using a Philips CM/12 TEM operated at 80 kV with images recorded on KODAK 4489 film.
Anatomical characterization
Anatomical characterization was carried out according to the IAWA standard (Wheeler and Gasson 1989). Briefly, one mean was calculated for each tree for each measurement to minimize pseudo replication. These means were based on 300 measurements for fiber lumen area, 100 for cell wall thickness, and a minimum of 60 for the remaining measurements. Fiber lumen area measurements were increased from the IAWA standard of 100 to 300 to account for longitudinal geometrical effects. One complete set of images were taken for the full area of each sector from cambium to pith. Morphological measurements were taken on fully differentiated cells, approximately 1 mm from the cambium. Image analysis was performed with ImageJ (Abramoff et al. 2004). Images of macerated fibers were taken of free fully hydrated fibers. Anatomical properties characterized were fiber length, width, lumen area, and double fiber cell wall thickness.
Statistical model for analysis
A multivariate statistical model was applied and evaluated in accordance to the methods outlined in Sandquist et al. (2015). The seven variables included in the multivariate model correspond to the measurements given in Table 3, excluding results for guaiacyl lignin. To improve estimates of variance, the model utilized the variance from the equivalent measurements given in Sandquist et al.(2015). As there were no equivalents for the UV-measurements of guaiacyl lignin, the guaiacyl measurements were only evaluated with a univariate version of the model composed of measurements from this study. The multivariate model used for the analysis was:
where n is the number of dimensions/variables, the vector of overall means,
the vector of genotype effects, and
are the fixed effects (genotypes in group A, B and C, respectively). The vectors of random effects for line and residual variation
are assumed to follow multivariate normal distributions with mean vectors 0 and covariance matrices:
The vectors and
are assumed independent.
RESULTS AND DISCUSSION
Group and Line Effects
Corresponding to previous studies (Sandquist et al. 2015), each genetic transformation was carried out twice in two independent lines. Trees were grown under similar conditions as described earlier (Sandquist et al. 2015). To acquire better estimates of variance, measurements in the present study were evaluated together with those previously reported (Sandquist et al. 2015) as an additional (third) group.
No statistically significant line variations were found over the extended population of three groups, except for fiber width (Table 2). Plots of line effects and residuals did not show any essential deviations from the assumption of normal distributions.
Overall Statistical Observations
Despite being down regulated in significant molecular pathways, we have been impressed with how well overall developed the three genotypes appear to be. Statistical relationships over the extended population of three- instead of two groups showed only minor variations to the previously reported correlations (Table 2) (Sandquist et al. 2015). The only differences were the indication of statistical significant relationships between fiber lumen area and plant diameter, and between plant height and fiber length. From previously reported tree growth patterns (Kollmann 1968) these indications are in accordance with expected correlations.
The main reason for utilizing estimates of variation from previous measurements was to improve the error estimation. Evaluation of the new population on its own (with 24 samples divided into 4 groups with two lines and three replicates) generates results that are over sensitive to line variations. This effect was observed in the ultraviolet guaiacyl lignin evaluation, which had no previous equivalent measurements. If line variations are neglected, however, the statistical model is reduced to genotypes with six replicates, and then genotypes KR112 and KR139 show a statistical significant decrease in UV-absorption (p-values ≤0.0001 and 0.0001, respectively).
Morphological Effects of Sucrose Synthase Down-regulation
Genotype KR112 with a relative reduction in density due to knock-down of the sucrose synthase gene (Gerber et al. 2014) showed reduced fiber development, with a strong overall significant difference compared to wild type (Table 3). This was mainly manifested in reduced fiber cell wall thickness (Fig. 1D and 2A vs. 2D), and subsequently increased fiber lumen area (Table 3). There were also indications of reduced phloroglucinol staining across the fiber – middle lamella – fiber cell wall compared to the vessels, as shown in Figs. 1E vs. 1H. There were also indications of reduced UV-absorption across the fiber-fiber cell walls (Table 3 and Fig. 1) although this was more difficult to discern visually in the UV-images (Figs. 1M vs. 1P). However, the Mäule staining remained unchanged (Figs. 1I vs. 1L), and intensity of staining at the ultrastructural level did not appear to be affected (Figs. 2A vs. 2D). The Mäule reaction is considered to be selective towards syringyl-lignin (Chaffey 2002), and this may therefore indicate a shift in the S/G ratio for genotype KR112, with a possible reduction of guaiacyl content. This observation corroborates previously reported observations in a shift in the S/G ratio (Gerber et al. 2014). Furthermore, the reported shrinkage upon drying (Gerber et al. 2014) has been observed to an even greater extent in these samples due to the embedding procedure, which further corroborates the observed change in cell wall architecture.
Table 2. Estimated Variances of Morphological Residuals and Correlations
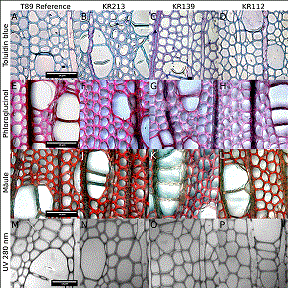
Fig. 1. Representative images of reference wildtype T89, genotypes KR213, KR139 and KR112. Staining was with toluidine blue (first row), phloroglucinol (second row) and Mäule reaction (third row). Bottom row shows UV-microscopy images at 280 nm. Scale is the same within each row. Top 3 rows show images at the same magnification to allow visual comparisons. The images reflect observations on a minimum of 3 sections, with multiple images per sample. Genotype KR213 shows no statistical difference compared with the reference, whereas KR112 shows reduced staining with phloroglucinol and genotypes KR139 shows reduced staining with both phloroglucinol and the Mäule reaction.
Morphological Effects of Cinnamate 4-hydroxylase Down-regulation
Genotype KR139 with a relative reduction of lignin due to knock-down of the cinnamate 4-hydroxylase coding gene (Bjurhager et al. 2010), showed only an indication of difference in fiber morphology compared to the wild type (Table 3). A visual comparison can be made between Figs. 1A vs. 1C. The reduced fiber length was the only individually significant observation (Table 3). Less intense staining was observed for both phloroglucinol and the Mäule reaction, as seen in Figs. 1E vs.1G, and 1I vs. 1K. There was also an indication of reduced lignin absorption in UV in the fiber-fiber cell wall (Fig. 1 and 1M vs. 1O) which was also reflected at the ultrastructural level (Figs. 2A vs. 2C). Together, this suggests normal wood development, with an overall reduction of lignin concentration in the fiber-fiber cell wall, including the middle lamella region. No shift in S/G ratio can be inferred. These observation confirm previously reported observations by Bjurhager et al. (2010) in the studied aspen genotype, Sewalt et al. (1997) in tobacco, and Reddy et al. (2005) in alfalfa. This further emphasizes the key role played by this enzyme in lignin biosynthesis.
Morphological Effects of Pectate Lyase Coding Gene Down-regulation
Genotype KR213 was previously reported with reduced fiber development (Sandquist et al. 2015), but in this extended study it did not show any overall significant differences. The previously reported results reflected aspen grown under summer conditions, whereas the present results reflect spring conditions (Sandquist et al. 2015). Despite having been grown under otherwise similar conditions in the green house, these results may indicate a difference between spring and summer development. Alternatively, this change could be caused by more accurate interpretation from a larger statistical population.
Even though the trends remain the same, the previously reported individual indications of reduced fiber lumen area and increased fiber cell wall thicknesses (Sandquist et al. 2015) were absent in the spring developing fibers (Figs. 1A vs. 1B). The earlier reported fiber length reduction are now significant (Table 1). No change in lignin composition were observed with the two lignin stains (Figs. 1E vs. 1F and 1I vs. 1J), UV-microscopy (Fig. 1, 1M vs. N) nor at the ultrastructural level (Fig. 2A vs.2B).
Despite being grown in a greenhouse under the same climate and (compensated) light conditions, an overall significant increase was observed in fiber lumen area from spring to summer compared to what was observed for the wild type. In addition, we noted a significant decrease in overall fiber length and width for the same period.
Ultrastructural TEM Observations
At the ultrastructural level, no major differences were observed in the secondary cell wall architecture of fibers and vessels (i.e. S1, S2) in any of the three genotypes in comparison to the reference plants using TEM at the magnification used. This contrasts with previous studies, in which changes in the secondary wall, particularly the S2 layer (e.g. concentric loosening of S2 wall layer), has been reported in poplar (Leple et al. 2007), Arabidopsis (Ruel et al. 2009), and flax (Day et al. 2009) in knock-down mutants affecting enzymes of lignin biosynthesis. The TEM observations were however consistent with the histochemical and UV-observations of fibers and vessel wall architecture, suggesting that any changes were likely to be on a supermolecular rather than ultrastructural level. The reduced UV-absorption for lignin at 280 nm of the double fiber wall (Reference > KR213 > KR139 > KR112, Table 3) was also consistent with TEM staining where the transgenic trees showed less staining of the fiber and vessel S2 and middle lamella regions using OsO4 and KMnO4 (Fig. 2).
Table 3. Summary of Properties
| Construct | Fiber
length |
Fiber
width |
Fiber lumen
area |
Radial fiber
thickness |
Tangential fiber
thickness |
| mm | µm | µm2 | µm | µm | |
| Ctrl | 1.03 | 38.5 | 36.7 | 2.0 | 1.8 |
| KR112 | 1.02 | 39.1 | 51.9 | 1.0 | 1.1 |
| KR139 | 0.97 | 38.8 | 40.0 | 1.7 | 1.8 |
| KR213 | 0.98 | 37.2 | 33.6 | 2.2 | 2.2 |
| Mean | 1.00 | 38.4 | 40.5 | 1.7 | 1.8 |
| Literature | 34.9b | 2.06a,2.01b | 2.00a,1.94b | ||
| Construct | Overall
multivariate analysis |
Plant
height |
Plant
diameter |
UV Guaiacyl | |
| p-values | cm | mm | max. intensity | ||
| Ctrl | – | 105.0 | 7.5 | 38500 | |
| KR112 | <0.001 | 105.0 | 7.6 | 30100 | |
| KR139 | 0.025 | 90.2 | 7.4 | 30900 | |
| KR213 | 0.056 | 92.3 | 7.0 | 35900 | |
| Mean | – | 98.2 | 7.4 | 33900 | |
| Bold underlined values are statistically significant at p ≤ 0.05 compared with control. Bold values show a statistical tendency of significance with p ≤ 0.10 compared with control. P-values are adjusted for multiple comparisons within groups. a(Roach et al. 2012). b(Sandquist et al. 2015). | |||||
Fig. 2. TEM images showing reference wildtype T89 (A), KR213 (B), KR139 (C) and KR112 (D). Samples were post-stained with OsO4 and sections post-stained with KMnO4. They are representative selections from multiple images per sample and line. The images confirm previous observations with no major morphological changes except for the apparent reduction in cell wall thickness in genotype KR112. Genotype KR139 shows less intense staining of the primary cell wall and particularly of the middle lamella cell corners. This corroborates both the UV and histochemical observations. Abbreviations: S1, S2, secondary cell wall layers; ML, middle lamella; MLcc, middle lamella cell corner; CL, cell lumen.
CONCLUSIONS
- Knock-down of the sucrose synthase (SUS4) coding gene in aspen significantly decreased fiber development. There were also indications that this decrease affects the S/G lignin ratio.
- Knock-down of the cinnamate 4-hydroxylase coding gene showed from the multivariate evaluation only an overall indication of modified fiber morphology. Chemical staining and UV-microscopy measurements indicate an overall reduction of lignin in the fibers, with no shift in S/G ratio.
- Knock-down of the pectate lyase family coding gene did not contribute to any overall observed changes in fiber morphology, nor lignin content.
ACKNOWLEDGMENTS
The authors gratefully acknowledge funding provided by the Formas FuncFiber Center of Excellence (www.funcfiber.se). The authors gratefully acknowledge Prof. Björn Sundberg for supplying the hybrid plants. We also want to thank Prof. Sundberg and SweTree Technologies (swetree.com) for the initial screening of these genotypes.
REFERENCES CITED
Abramoff, M., Magelhaes, P., and Ram, S. (2004). “Image processing with ImageJ,” Biophotonics International 11(7), 36-42.
Baucher, M., Halpin, C., Petit-Conil, M., and Boerjan, W. (2003). “Lignin: Genetic engineering and impact on pulping,” Critical Reviews in Biochemistry and Molecular Biotechnology 38, 305-350. DOI: 10.1080/10409230390242443
Bjurhager, I., Olsson, A. M., Zhang, B., Gerber, L., Kumar, M., Berglund, L., Burgert, I., Sundberg, B., and Salmén, L. (2010). “Ultrastructure and mechanical properties of Populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase,” Biomacromolecules11, 2359-2365. DOI: 10.1021/bm100487e
Boerjan, W., Ralph, J., and Baucher, M. (2003). “Lignin biosynthesis,” Annual Review of Plant Biology 54, 519-546. DOI: 10.1146/annurev.arplant.54.031902.134938
Chaffey, N. L. (2002). “Wood microcopy techniques” in: Wood Formation in Trees, N.L. Chaffey (ed), Taylor & Francis Inc, London. DOI: 10.1093/aob/mcf216
Chiang, V. (2002). “From rags to riches,” Nature Biotechnology 20, 557-558. DOI:10.1038/nbt0602-557
Day, A., Neutelings, G., Nolin, F., Grec, S., Habrant, A., Cronier, D., Maher, B., Rolando, C., David, H., Chabbert, B., and Hawkins, S. (2009). “Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem,” Plant Physiology and Biochemistry 47, 9-19. DOI: 10.1016/j.plaphy.2008.09.011
Fengel, D., and Wegener, G. (1989). “3. Chemical composition and analysis of wood,” in: Wood: Chemistry, Ultrastructure, Reactions, Walter De Gruyter, Berlin. DOI: 10.1515/9783110839654
Gerber, L., Zhang, B., Roach, M., Rende, U., Gorzsás, A., Kumar, M., Burgert, I., Niittylä, T., and Sundberg, B. (2014). “Deficient sucrose synthase activity in developing wood does not specifically affect cellulose biosynthesis, but causes an overall decrease in cell wall polymers,” New Phytologist203(4), 1220-1230. DOI: 10.1111/nph.12888
Horvath, B., Peszlen, I., Peralta, P., Kasal, B., and Li, L. (2010). “Effect of lignin genetic modification on wood anatomy of aspen trees,” IAWA Journal 31(1), 29-38. DOI: 10.1163/22941932-90000003
Hu, W., Harding, S., Lung, J., Popko, L., Ralph, J., Stokke, D., Tsai, C., and Chiang, V. (1999). “Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees,” Nature Biotechnology 17, 808-812. DOI:10.1038/11758
Karimi, M., Inze, D., and Depicker, A. (2002). “GATEWAY™ vectors for Agrobacterium-mediated plant transformation,” Trends in Plant Science 7(5), 193-195. DOI: 10.1016/S1360-1385(02)02251-3
Kollmann, F. (1968). “6. Physics of wood,” in: Principles of Wood Science and Technology, Vol. 1, Kollmann, F., and Côté W. J. (eds.), Springer Verlag, Berlin. ISBN 978-3-642-87928-9
Leple, J. C., Dauwe, R., Morreel, K., Storme, V., Lapierre, C., Pollet, B., Naumann, A., Kang, K. Y., Kim, H., Ruel, K., Lefebvre, A., Joseleau, J. P., Grima-Pettenati, J., Rycke, R., Andersson-Gunneras, S., Fehrle, I., Petit-Conil, M., Kopka, J., Polle, A., Messens, E., Erban, A., Sundberg, B., Mansfield, S., Ralph, J., Pilate, G., and Boerjan, W. (2007). “Downregulation of cinnamoyl-coenzyme a reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure,” The Plant Cell 19, 3669-3691. DOI: 10.1105/tpc.107.054148
Li, L., Zhou, Y., Cheng, X., Sun, J., Marita, J., Ralph, J., and Chiang, V. (2003). “Combinatorial modification of multiple lignin traits in trees through multigene cotransformation,” PNAS 100(8), 4939-4944. DOI: 10.1073/pnas.0831166100
Nilsson, O., Aldén, T., Sitbon, F., Little, A. C. H., Chalupa, V., Sandberg, G., and Olsson, O. (1992). “Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging,” Transgenic Research 1, 209-220. DOI: 10.1007/BF02524751
Pilate, G., Guiney, E., Holt, K., Petit-Conil, M., Lapierre, C., Leplé, J.C., Pollet, B., Mila, I., Webster, E., Marstorp, H., Hopkins, D., Jouanin, L., Boerjan, W., Schuch, W., Cornu, D., and Halpin, C. (2002). “Field and pulping performances of transgenic trees with altered lignification,” Nature Biotechnology20, 607-612. DOI: 10.1038/nbt0602-607
Reddy, M. S. S., Chen, F., Shadle, G., Jackson, L., Aljoe, H., and Dixon, R. A. (2005). “Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.),” PNAS 102, 16573-16578. DOI: 10.1073pnas.0505749102
Roach, M., Gerber, L., Sandquist, D., Gorzsás, A., Hedenström, M., Kumar, M., Steinhauser, M. C., Feil, R., Daniel, G., Stitt, M., Sundberg, B., and Niittylä, T. (2012). “Fructokinase is required for carbon partitioning to cellulose in aspen wood,” Plant Journal 70(6), 967-977. DOI: 10.1111/j.1365-313X.2012.04929.x
Ruel, K., Berrio-Sierra, J., Derikvand, M. M., Pollet, B., Thevenin, J., Lapierre, C., Jouanin, L., and Joseleau, J. P. (2009). “Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana,” New Phytologist 184, 99-113. DOI: 10.1111/j.1469-8137.2009.02951.x
Sandquist, D., Norell, L., and Daniel, G., (2015). “Quantitative evaluation of hybrid aspen xylem and immunolabeling patterns using image analysis and multivariate statistics,” BioResources 10(3), 4997-5015.
Sewalt, V. J. H., Ni, W., Blount, J. W., Jung, H. G., Masoud, S. A., Howles, P. A., Lamb, C., and Dixon, R. A. (1997). “Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase,” Plant Physiology 115, 41-50. DOI: 10.1104/pp.115.1.41
Sjödin, A., Street, N. R., Sandberg, G., Gustafsson, P., and Jansson, S. (2009). “The Populus genome integrative explorer (PopGenIE): A new resource for exploring the Populus genome,” The New Phytologist 182(4), 1013-1025. DOI:10.1111/j.1469-8137.2009.02807.x. PMID 19383103
Sjöström, E. (1993). “The structure of wood,” in: Wood Chemistry – Fundamentals and Applications, 2nd Ed., Academic Press Inc., London. ISBN 0-12-647481-8
Spurr, A. R. (1969). “A low-viscosity epoxy resin embedding medium for electron microscopy,” Journal of Ultrastructure Research 26(1-2), 31-43. DOI: 10.1016/S0022-5320(69)90033-1
Tuskan, G., DiFazio, S., and Teichmann, T. (2003). “Poplar genomics is getting popular – The impact of the poplar genome project on tree research,” Plant Biology 5, 1-3. DOI: 10.1055/s-2003-44715
Vanholme, R., Morreel, K., Ralph, J., and Boerjan, W. (2008). “Lignin engineering,” Curr. Opin. Plant Biol. 11(3), 1-8. DOI: 10.1016/j.pbi.2008.03.005
Wheeler, E., and Gasson, P. (eds.) (1989). “IAWA list of microscopic features for hardwood identification,” International Association of Wood Anatomists (IAWA), Reprinted from IAWA Bulletin n.s. 10(3), 219-332.
Wise, L. E., and Murphy, M. (1946). “A chlorite holocellulose, its fractionation and bearing on summative wood analysis and studies on the hemicelluloses,” Paper Trade Journal 122, 35-43.
Yoshinaga, A., and Wada, M. (2007). “Modified lignification in the cell walls of cad depressed poplars,” IAWA Journal 28(4), 457-471. DOI: 10.1163/22941932-90001655
Article submitted: February 7, 2015; Peer review completed: March 28, 2015; Revisions accepted: June 9, 2015; Published: June 25, 2015.
DOI: 10.15376/biores.10.3.5016-5029
