Abstract
The heterologous expression of the gene LACP83 (encoding a laccase) from Pleurotus ostreatus in Escherichia coli was characterized. The laccase enzyme activity and kinetics of bacterial growth with an inducer (IPTG) and without inducer were determined. The maximum enzymatic activity was observed at 7 h post induction with a value of 3740 ± 342 U/L, which was similar to that reported for the native strain of P. ostreatus at 144 h of culture. Furthermore, the induction of laccase with IPTG reduced the specific growth rate of recombinant E. coli BL21 by approximately 50%. These results support the use this system for the recombinant production of the enzyme on an industrial scale.
Download PDF
Full Article
Heterologous Expression of Laccase (LACP83) of Pleurotus ostreatus
Amado I. Grandes-Blanco,a,c Saúl Tlecuitl-Beristain,c Rubén Díaz,b Carmen Sánchez,b Maura Téllez-Téllez,d Luis Márquez-Domínguez,e Gerardo Santos-López,e and Gerardo Díaz-Godínez b,*
The heterologous expression of the gene LACP83 (encoding a laccase) from Pleurotus ostreatus in Escherichia coli was characterized. The laccase enzyme activity and kinetics of bacterial growth with an inducer (IPTG) and without inducer were determined. The maximum enzymatic activity was observed at 7 h post induction with a value of 3740 ± 342 U/L, which was similar to that reported for the native strain of P. ostreatus at 144 h of culture. Furthermore, the induction of laccase with IPTG reduced the specific growth rate of recombinant E. coli BL21 by approximately 50%. These results support the use this system for the recombinant production of the enzyme on an industrial scale.
Keywords: Gene; Recombinant protein; Transformant
Contact information: a: Doctorado en Ciencias Biológicas, Universidad Autónoma de Tlaxcala, Tlaxcala, México; b: Laboratorio de Biotecnología, Centro de Investigacion en Ciencias Biológicas, Universidad Autónoma de Tlaxcala, Tlaxcala, México; c: Laboratorio de Biotecnología, Universidad Politécnica de Tlaxcala, San Pedro Xalcaltzinco, Tlaxcala, México; d: Centro de Investigaciones Biológicas, Universidad Autónoma del Estado de Morelos, Cuernavaca, Morelos, México; e: Centro de Investigación Biomédica de Oriente Instituto Mexicano del Seguro Social, Metepec, Puebla, México;
* Corresponding author: diazgdo@hotmail.com
INTRODUCTION
Laccases are enzymes found in plants (Wang et al. 2015), insects (Ye et al. 2015), bacteria (Tonin et al. 2016), and white rot fungi (Yang et al. 2015). These enzymes use copper as a cofactor and oxidize a wide range of phenolic compounds including mono-, di- and polyphenols, aminophenols, methoxyphenols, and aromatic amines, with the consequent reduction of molecular oxygen to water (Madhavi and Lele 2009); the oxidation of these compounds generates free radicals that polymerize or degrade (Claus 2003). Laccase isoenzymes have different physicochemical properties (Téllez-Téllez et al. 2012a). They are widely used in industry and biotechnology processes, such as the paper, textile, food, and pharmaceutical industries, and their applications have extended to the synthesis of compounds in nanobiotechnology (Barreca et al. 2003; Alper and Acar 2004; Nicotra et al. 2004; Gamelas et al. 2005; Pazarlıoǧlu et al. 2005; García-Rivero et al. 2015). Pleurotus ostreatus is a basidiomycete and a laccase producer. Its maximum enzyme activity is found in its stationary growth phase at about 360 h of culture (Téllez-Téllez et al. 2008); therefore, the production time of the enzyme is limiting. These enzymes are induced, and the transcription gene of laccases is regulated by different carbon and nitrogen sources (Bertrand et al. 2013). In strategies to increase production, inducers have been added to the culture medium of the fungus, such as 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) (Hou et al. 2004), ferulic acid (Yao et al. 2013), plant residues (Quevedo-Hidalgo et al. 2015), and azo dyes (Camacho et al. 2015). However, production times are still very long and costly.
Molecular tools have also been used to increase the production of laccase. There are over 1200 laccase gene sequences stored in the database of the European Molecular Biology Laboratory (EMBL) (https://www.embl.de). More than half of these genes belong to fungi, and several have been expressed in homologous and heterologous systems, for example, laccase genes from Schizophyllum commune in Aspergillus sojae (Hatamoto et al. 1999), from Trametes versicolor in Pichia pastoris (Hong et al. 2002), and from Trametes sp. in P. methanolica (Hong et al. 2007).
Laccases from P. ostreatus have only been expressed in eukaryotic systems such as Kluyveromyces lactis, Saccharomyces cerevisiae (Piscitelli et al. 2005), Trichoderma reesei (Dong et al. 2012), and P. pastoris (Zhang et al. 2005; Park et al. 2015; Rivera-Hoyos et al. 2015), and to date, there is no report on their expression in a prokaryotic system such as E. coli. This bacterium has been widely used for gene expression because it is genetically characterized, spreads easily in simple media with a high rate of cell growth, and can produce high levels of recombinant protein (Sugantha et al. 2010; Wang et al. 2014; Wurm et al. 2016); thus, it is an interesting alternative for the expression of laccases genes from fungi. In this study, a laccase gene from P. ostreatus called LacP83 (Téllez-Téllez et al. 2012b) was cloned and expressed in E. coli.
EXPERIMENTAL
Materials and Methods
Strains and culture media
Strains of E. coli TOP 10 and BL21 (Thermo Fisher Scientific, Waltham, MA, USA) were grown in Luria-Bertani (LB) medium and incubated at 37 °C for 18 h at 250 rpm. The culture medium used to select transformed cells was LB with 2 mg of ampicillin per mL (LB/Amp) and the same incubation conditions as above. The culture medium used for induction of the recombinant gene was LB/Amp added with 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) and 0.25 mM CuSO4, as reported by Guan et al. (2014). The culture media and the chemical compounds used in this step were supplied by Sigma-Aldrich (Toluca, Mexico).
Obtaining and amplification of the gene LacP83
The sequence of gene LacP83 was obtained from the GenBank database with accession number JF719064 and chemically synthesized (Integrated DNA Technologies, Coralville, IA, USA). For expression, the reading frame was amplified with forward (5’-CACGGATCCTCCAGGCGCACGGATCTCGCTACGCTTACA-3’) and reverse primers (5’-TAATTAAGCTTTCAAGCTATGCCACCTTTGTCGGAATCGCTC-3’) that have recognition sites for the restriction enzymes BamHI and HindIII (underlined in the sequence). The PCR conditions used were: 95 °C for 2 min, 30 cycles of 95 °C for 30 s, 56 °C for 50 s, 72 °C for 2 min, and a final extension at 72 °C for 5 min. In all cases, the genetic material obtained was corroborated by electrophoresis on a 0.8% agarose gel (Sambrook and Russell 2001) with a 1 kb molecular marker (Thermo Fisher Scientific, Waltham, MA, USA).
Cloning vector
The amplified DNA obtained from the above step was inserted into the cloning vector pJET 1.2 Blunt (Fermentas Thermo Fisher Scientific, Waltham, MA, USA) following the supplier’s instructions and cloned into E. coli TOP 10. Plasmid purification was performed by alkaline lysis (Sambrook and Russell 2001), and the desired insert was confirmed by a restriction profile.
Expression vector
The obtained DNA from restriction profile was subcloned into the expression vector pQE30 (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The resulting plasmid was introduced into the strain E. coli BL21 using an electroporator (Eppendorf Mod 2510, Hamburg, Germany). The transformed cells were selected, and the plasmid was purified by alkaline lysis (Sambrook and Russell 2001). The presence of the gene was verified by PCR under the conditions described above. The presence of the reading frame of LacP83 in the recombinant plasmid pQEP83 was confirmed by PCR, and after that, the recombinant strain of E. coli BL21 was grown.
Growth kinetics of recombinant strain of E. coli BL21
The recombinant strain of E. coli BL21 was grown in LB/Amp medium, and the expression of the gene LacP83 was induced by adding IPTG when the culture reached an optical density of 0.5 at 600 nm (approximately at 12 h of culture) (Sugantha et al. 2010). Samples were taken from the induction every hour until 35 h of cultivation, and the biomass was evaluated by optical density at 600 nm in a Cary 300 Bio UV-Vis spectrophotometer (Agilent Technologies, Santa Clara, CA, USA). The kinetic parameters of growth were estimated with the Velhurst-Pearl logistic equation using the non-linear least square-fitting program “Solver” (Excel software, Microsoft, Redmond, WA, USA) (Téllez-Téllez et al. 2008).
Enzymatic activity assay
Laccase activity was determined at 40 °C in each sample by changes in the absorbance at 468 nm, using 2 mM 2,6-dimethoxyphenol (DMP) in phosphate buffer pH 6.5 as the substrate (Téllez-Téllez et al. 2008). One enzymatic unit (U) of laccase activity was defined as the amount of enzyme that gives an increase of 1 unit of absorbance per min in the reaction mixture. The storage stability of the laccase was tested; the activity after 15 and 30 days storage at 4 °C was assessed. Protein patterns were observed via SDS-PAGE (Laemmli 1970). All chemical compounds for laccase activity and electrophoresis were supplied by BioRad (Hercules, California, USA).
RESULTS AND DISCUSSION
The construction of expression vector for E. coli (pQEP83) and the PCR product are shown in Figs. 1 and 2, respectively. A fragment of approximately 1599 bp was observed, as expected. The characteristics bands of extraction from plasmid DNA of the expression vector called pQE30P83 in E. coli BL21 and PCR product are shown in Fig. 3. A fragment of approximately 1600 bp was observed, which confirmed that the purified plasmid contained the gene LacP83.
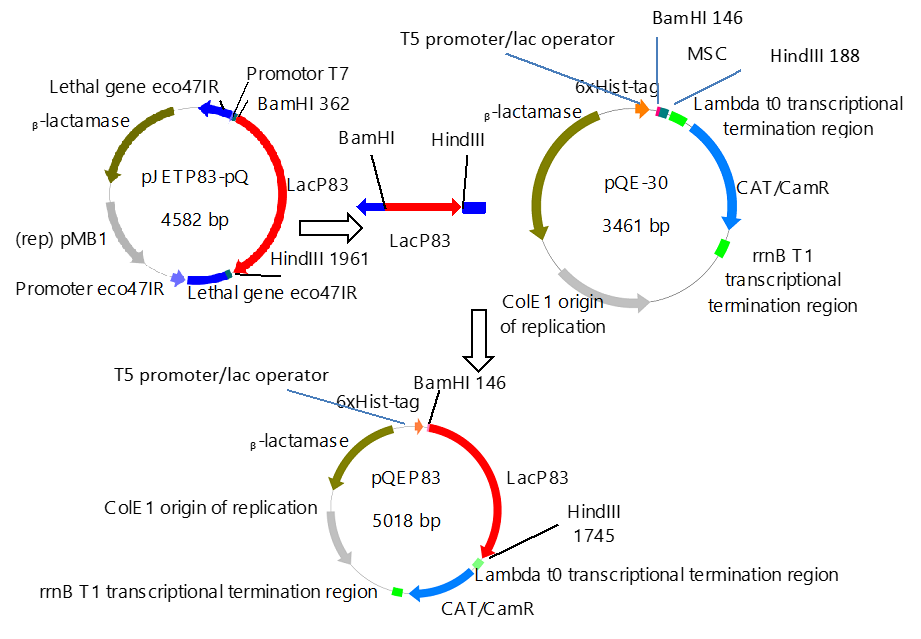
Fig. 1. Construction of the expression vector pQEP83
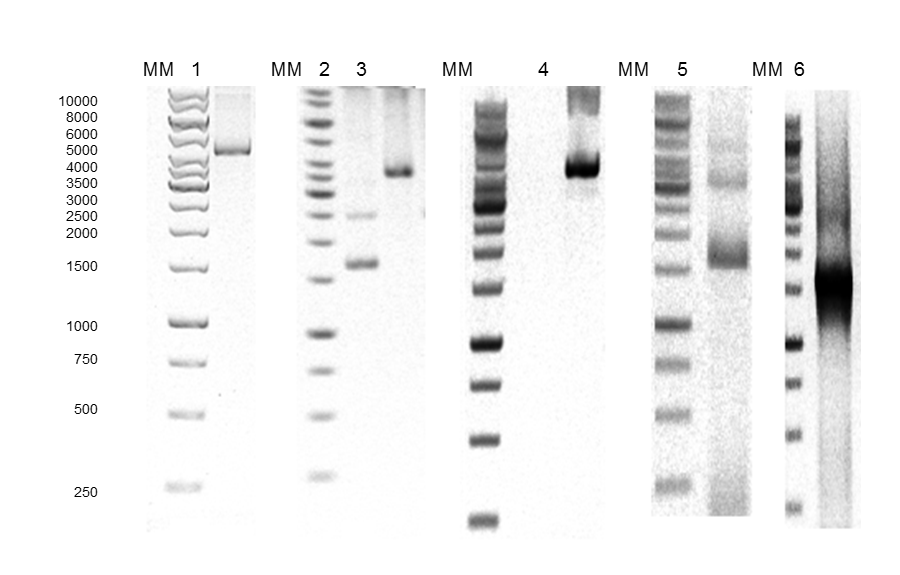
Fig. 2. Agarose gel electrophoresis. A) 1: pJETP83 after cutting with HindIII, 2: pJETP83 after cutting with HindIII and BamHI, 3: pQE30 after cutting with BamHI and HindIII, 4: pQEP83 after cutting with HindIII, 5: pQEP83 after cutting with HindIII and BamHI, 6: PCR product of pQEP83, MM: molecular marker 1 Kb
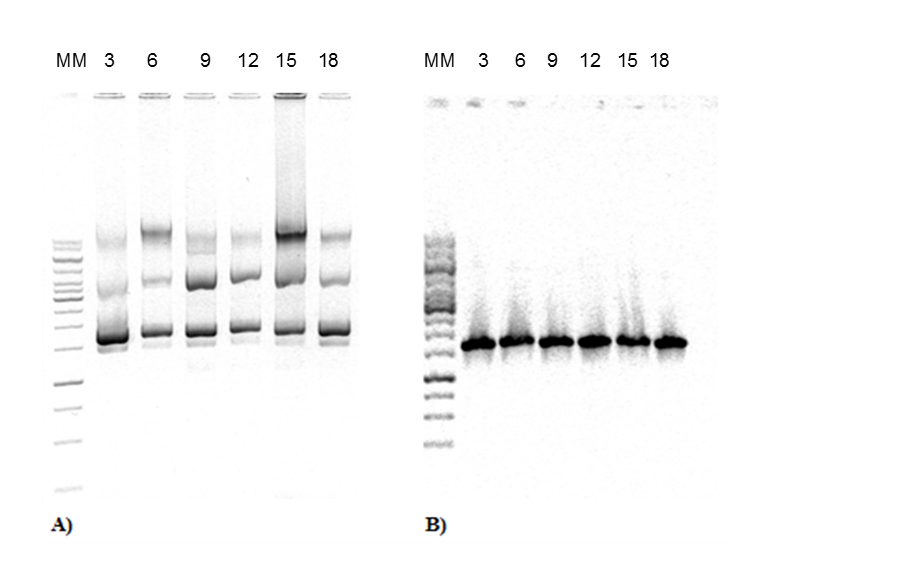
Fig. 3. DNA extraction and PCR product from expression strain. A) Purification of plasmid DNA from E. coli BL21 with recombinant pQEP83 at 3-18 h of culture. B) PCR of plasmids from E. coli BL21 with recombinant plasmid pQEP83. The number represents the time of sampling. MM: molecular marker 1 Kb
The LacP83 gene encodes for a constitutive laccase in P. ostreatus ATCC-32783 (Téllez-Téllez et al. 2008, 2012) with good catalytic characteristics, showing high stability a changes of pH and temperature in addition, the promoter of the gene presents sequences of response to stress, xenobiotics and metals. However, its production in the wild strain requires more than 20 days so a heterologous system where it is possible to increase its production in less time represents a good option for later studies.
Figure 4 shows the growth of E. coli BL21 recombinant (without induction and with induction). The exponential phase began approximately 7 h after inoculation. However, growth in the presence of IPTG was slower, although the maximum biomass was similar in both cultures after 35 h. In the absence of IPTG, the stationary phase occurred at approximately 18 h of culture, whereas in the presence of IPTG, it was close to 30 h of culture. The specific growth rate (ᶙ) for culture without IPTG was 0.536 h-1 and with IPTG was reduced by approximately 50% (0.244 h-1). A similar result was reported by Wagner et al. (2008), who found a decrease in the growth of E. coli BL21 after the addition of IPTG for inducing a recombinant protein. Vilar et al. (2003) argued that the decline in growth of recombinant strains may be due to changes of induction of lac promoter that affect cell growth. This effect can be inhibited by adding xylose the culture medium as an alternative carbon source (Gálvez et al. 2014).
The activity of the recombinant laccase (LacP83) is shown in Fig. 5. Two activity peaks were observed. The first (1800 U/L) occurred approximately 1 h after induction, and the second (3740 U/L), which was higher, was observed 6 h after the first. Ihssen et al. (2015) reported an activity of 3400 U/L of a laccase obtained from Bacillus licheniformis and expressed in E. coli. It is noteworthy that the laccase activity observed in the present study was 80 times higher than the laccase activity of Aeromonas hydrophila NIU01 expressed in E. coli (Ng et al. 2013). In addition, the value of laccase activity of this work was 2 times higher than that reported by Park et al. (2015), where the Lacc6 gene from P. ostreatus was expressed in P. pastoris, obtaining values of 1560 U/L. The wild strain of P. ostreatus produced 3190 U/L at 144 h of culture (Grandes-Blanco et al. 2013), while in this work, the recombinant strain of E. coli produced a similar activity in 19 h.
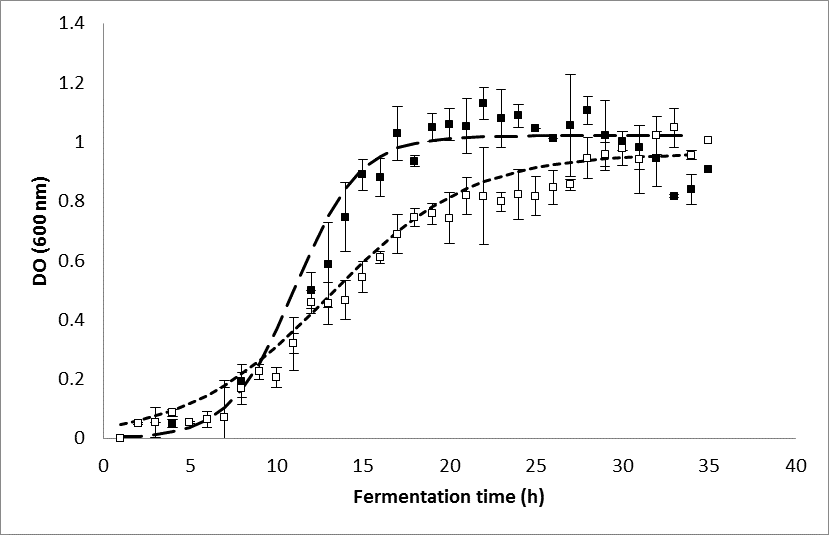
Fig. 4. Cellular growth (OD600) of recombinant E. coli BL21. (▪) Biomass without induction; (▫) biomass with induction. The vertical dotted line represents the time of induction with IPTG.
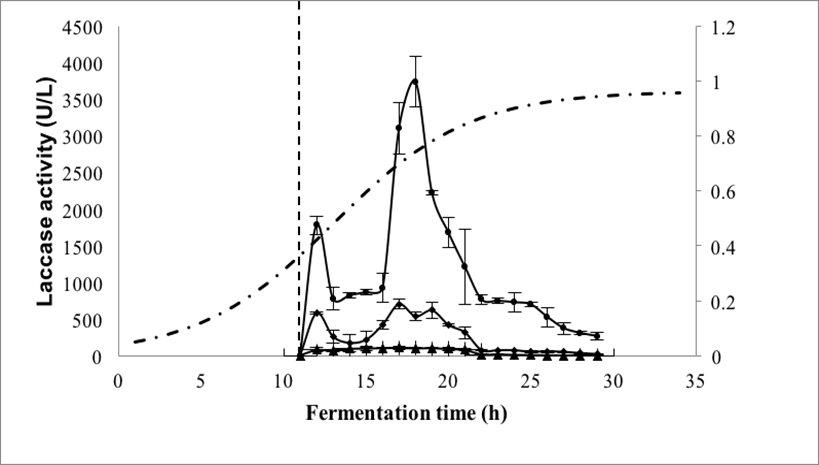
Fig. 5. Laccase activity from recombinant E. coli BL21 added with IPTG. (●) Immediately after sampling; (▪) after 15 days of storage; ◊ after 30 days of storage. (·-·) Cellular growth. The vertical dotted line represents the time of induction with IPTG.
Laccase activity was degraded by storage, losing approximately 90% in 15 days and almost 100% in 30 days at 4 °C. Gunne et al. (2013) suggested that the intracellular accumulation of copper does not allow adequate expression of the protein and that E. coli BL21 does not perform posttranslational modifications efficiently. It is noteworthy that despite this, E. coli grows 16 times faster (μ = 0.536 h-1) than P. ostreatus ATCC-32783 (μ = 0.032 h-1). On the other hand, Alexandre and Zhulin (2000) reported that E. coli produces proteins (PcoA_1073341 and YacK_2506227) with high laccase similarity and Outten et al. (2001) reported that this bacterium presents mechanisms of response to the increase of copper during its growth, which produce multicopper oxidases that are very similar to the laccases. These enzymes do not need to be glycosylated to be functional, which led us to suppose that the non-glycosylated laccases would have activity and stability. One option to reduce this problem would be the co-expression of intracellular chaperone proteins.
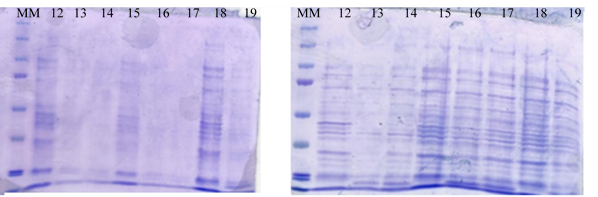
The protein expression profile of recombinant E. coli culture BL21 is shown in Fig. 6. The intensity of the bands increased from 5 h and in the exponential phase when the inducer (IPTG) was added, indicating that the induction of protein expression with IPTG to express laccase also stabilized other proteins. Leone et al. (2015) mentioned that in the recombinant system of E. coli BL21, the greatest expression of the protein occurs during the stationary phase even when IPTG was added. In the present study, protein expression was higher in the exponential phase of microbial growth.
A B
Fig. 6. SDS-PAGE of the culture medium during the growth of recombinant E. coli BL21. The number represent the time of sampling. A) Without induction. B) With induction (IPTG). MM: molecular marker.
CONCLUSIONS
- A recombinant plasmid (pQE30P83) was generated for transformation into coli BL21, from the LacP83 gene, which expresses a laccase of P. ostreatus ATCC-32783 with high activity and stability at changes in pH and temperature.
- The LacP83 gene expression was induced with IPTG. The laccase secreted to the medium had an enzyme activity similar to or greater than that reported in other homologous systems.
- A similar activity was observed to that reported for the native strain of ostreatus but in less time (86% less). A reduction of production time is desirable for the bioprocess because it reduces production costs.
- Laccase activity decreased with time and storage conditions, possibly by instability due to lack of glycosylation, so posttranslational modifications may be necessary to stabilize the enzyme.
ACKNOWLEDGMENTS
The authors thank the Mexican Council of Science and Technology (CONACyT) for supporting this research with Project No. 156406 and for the scholarship (No. 241720) awarded to Grandes-Blanco.
REFERENCES CITED
Alexandre, G., and Zhulin, I. B. (2000). “Laccases are widespread in bacteria,” Trends in biotechnology 18(2), 41-42. DOI: 10.1016/S0167-7799(99)01406-7
Alper, N., and Acar, J. (2004). “Removal of phenolic compounds in pomegranate juices using ultrafiltration and laccase‐ultrafiltration combinations,” Food/Nahrung 48(3), 184-187. DOI: 10.1002/food.200200258
Barreca, A. M., Fabbrini, M., Galli, C., Gentili, P., and Ljunggren, S. (2003). “Laccase/mediated oxidation of a lignin model for improved delignification procedures,” Journal of Molecular Catalysis B: Enzymatic 26(1), 105-110.
Bertrand, B., Martínez-Morales, F., and Trejo-Hernández, M. R. (2013). “Lacasas fungales: Inducción y producción,” Revista Mexicana de Ingeniería Química 12(3), 473-488.
Camacho, J., Nava, B., Díaz, R., Tlecuitl-Beristain, S., Garrido, V., and Bibbins-Martínez, M. (2015). “Induction effect of azo yellow dye on gene expression and activity of oxidases of Pleurotus ostreatus grown in submerged fermentation,” Revista Latinoamericana el Ambiente y las Ciencias 6, 57-71.
Claus, H. (2003). “Laccases and their occurrence in prokaryotes,” Archives of Microbiology 179(3), 145-150. DOI: 10.1007/s00203-002-0510-7.
Dong, X., Qin, L., Tao, Y., Huang, J., and Dong, Z. (2012). “Overexpression and characterization of a laccase gene from Pleurotus ostreatus in Trichoderma reesei,” Wei sheng wu xue bao= Acta microbiologica Sinica 52(7), 850-856.
European Molecular Biology Laboratory (EMBL) (2016). (https://www.embl.de), Accessed 20 November 2016.
Gamelas, J. A., Tavares, A. P., Evtuguin, D. V., and Xavier, A. M. (2005). “Oxygen bleaching of kraft pulp with polyoxometalates and laccase applying a novel multi-stage process,” Journal of Molecular Catalysis B: Enzymatic 33(3), 57-64. DOI: 10.1016/j.molcatb.2005.03.001
García-Rivero, M., Membrillo-Venegas, I., Vigueras-Carmona, S. E., Zafra-Jiménez, G., Zárate-Segura, P. B., and Martínez-Trujillo, M. A. (2015). “Enzymatic pretreatment to enhance chemical bleaching of akraft pulp,” Revista Mexicana de Ingeniería Química 14(2), 335-345.
Grandes-Blanco, A. I., Díaz-Godínez, G., Téllez-Téllez, M., Delgado-Macuil, R. J., Rojas-López, M., and Bibbins-Martínez, M. D. (2013). “Ligninolytic activity patterns of Pleurotus ostreatus obtained by submerged fermentation in presence of 2, 6-dimethoxyphenol and remazol brilliant blue R dye,” Journal Preparative Biochemistry and Biotechnology 43(5), 468-480. DOI: 10.1080/10826068.2012.746233.
Guan, Z. B., Song, C. M., Zhang, N., Zhou, W., Xu, C. W., Zhou, L. X., Zhao, H., Cai, Y. J., and Liao, X. R. (2014). “Overexpression, characterization, and dye-decolorizing ability of a thermostable, pH-stable, and organic solvent-tolerant laccase from Bacillus pumilus W3,” Journal of Molecular Catalysis B: Enzymatic 101, 1-6. DOI: 10.1016/j.molcatb.2013.11.009
Gunne, M., Al-Sultani, D., and Urlacher, V. B. (2013). “Enhancement of copper content and specific activity of CotA laccase from Bacillus licheniformis by coexpression with CopZ copper chaperone in E. coli,” Journal of Biotechnology 168(3), 252-255. DOI: 10.1016/j.jbiotec.2013.06.011
Hatamoto, O., Sekine, H., Nakano, E., and Abe, K. (1999). “Cloning and expression of a cDNA encoding the laccase from Schizophyllum commune,” Bioscience, Biotechnology, and Biochemistry 63(1), 58-64. DOI:10.1271/bbb.63.58
Hong, F., Meinander, N. Q., and Jönsson, L. J. (2002). “Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris,” Biotechnology and Bioengineering 79(4), 438-449. DOI: 10.1002/bit.10297
Hong, Y. Z., Zhou, H. M., Tu, X. M., Li, J. F., and Xiao, Y. Z. (2007). “Cloning of a laccase gene from a novel basidiomycete Trametes sp. 420 and its heterologous expression in Pichia pastoris,” Current Microbiology 54(4), 260-265. DOI: 10.1007/s00284-006-0068-8
Hou, H., Zhou, J., Wang, J., Du, C., and Yan, B. (2004). “Enhancement of laccase production by Pleurotus ostreatus and its use for the decolorization of anthraquinone dye,” Process Biochemistry 39(11), 1415-1419. DOI: 10.1016/S0032-9592(03)00267-X
Ihssen, J., Reiss, R., Luchsinger, R., Thöny-Meyer, L., and Richter, M. (2015). “Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in Escherichia coli,” Scientific Reports 5, 10465. DOI:10.1038/srep10465
Laemmli, U. K. (1970). “Cleavage of structural proteins during the assembly of the head of bacteriophage T4,” Nature 227, 680-685. DOI: 10.1038/227680a0
Leone, S., Sannino, F., Tutino, M. L., Parrilli, E., and Picone, D. (2015). “Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source,” Microbial Cell Factories 14(1), 1. DOI: 10.1186/s12934-015-0299-0
Madhavi, V., and Lele, S. S. (2009). “Laccase: Properties and applications,” BioResources 4(4), 1694-1717.
Ng, I. S., Zhang, X., Zhang, Y., and Lu, Y. (2013). “Molecular cloning and heterologous expression of laccase from Aeromonas hydrophila NIU01 in Escherichia coli with parameters optimization in production,” Applied Biochemistry and Biotechnology 169(7), 2223-2235. DOI: 10.1007/s12010-013-0128-z
Nicotra, S., Cramarossa, M. R., Mucci, A., Pagnoni, U. M., Riva, S., and Forti, L. (2004). “Biotransformation of resveratrol: Synthesis of trans-dehydrodimers catalyzed by laccases from Myceliophtora thermophyla and from Trametes pubescens,” Tetrahedron 60(3), 595-600. DOI: 10.1016/j.tet.2003.10.117
Outten, F. W., Huffman, D. L., Hale, J. A., and O’Halloran, T. V. (2001). “The independent cue and cusSystems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli,” Journal of Biological Chemistry 276(33), 30670-30677. DOI 10.1074/jbc.M104122200
Park, M., Kim, M., Kim, S., Ha, B., and Ro, H. S. (2015). “Differential expression of laccase genes in Pleurotus ostreatus and biochemical characterization of laccase isozymes produced in Pichia pastoris,” Mycobiology 43(3), 280-287. DOI: 10.5941/MYCO.2015.43.3.280.
Pazarlıoǧlu, N. K., Sariişik, M., and Telefoncu, A. (2005). “Laccase: Production by Trametes versicolor and application to denim washing,” Process Biochemistry 40(5), 1673-1678. DOI: 10.1016/j.procbio.2004.06.052.
Piscitelli, A., Giardina, P., Mazzoni, C., and Sannia, G. (2005). “Recombinant expression of Pleurotus ostreatus laccases in Kluyveromyces lactis and Saccharomyces cerevisiae,” Applied Microbiology and Biotechnology 69(4), 428-439. DOI: 10.1007/s00253-005-0004-z.
Quevedo-Hidalgo, B., Pedroza-Rodríguez, A. M., and Velásquez-Lozano, M. E. (2015). “Production of lignocellulolytic enzymes from floriculture residues using Pleurotus ostreatus,” Universitas Scientiarum 20(1), 117-127. DOI: 10.11144/Javeriana.SC20-1.eple
Rivera-Hoyos, C. M., Morales-Álvarez, E. D., Poveda-Cuevas, S. A., Reyes-Guzmán, E. A., Poutou-Piñales, R. A., Reyes-Montaño, E. A., Pedroza-Rodríguez, A. M., Rodríguez-Vazquez R., and Cardozo-Bernal, Á. M. (2015). “Computational analysis and low-scale constitutive expression of laccases synthetic genes GlLCC1 from Ganoderma lucidum and POXA 1B from Pleurotus ostreatus in Pichia pastoris,” PLOS ONE 10(1), e0116524. DOI: 10.1371/journal.pone.0116524
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual (3rd Ed.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA.
Sugantha , P., Gowri, S., Thirumalaisamy, R., Kavitha, P., Prakash, B., Arunachalam, G., and Selvamuthukumar, S. (2010). “Over expression of IPTG inducible GST protein in E. coli BL21,” Journal Biomedical Science and Research 2, 54-59.
Téllez-Téllez, M., Sánchez, C., Díaz, R., and Díaz-Godínez, G. (2012a). “Zymogram patterns of extracellular laccases of Pleurotus species grown on non-inducer agar medium,” Revista Mexicana de Ingeniería Química 11(3), 383-388.
Téllez-Téllez, M., Diaz-Godinez, G., Aguilar, M. B., Sánchez, C., and Fernández, F. J. (2012). “Description of a laccase gene from Pleurotus ostreatus expressed under submerged fermentation conditions,” BioResources 7(2), 2038-2050. DOI: 10.15376/biores.7.2.2038-2050
Téllez-Téllez, M., Fernández, F. J., Montiel-González, A. M., Sánchez, C., and Díaz-Godínez, G. (2008). “Growth and laccase production by Pleurotus ostreatus in submerged and solid-state fermentation,” Applied Microbiology and Biotechnology 81(4), 675-679. DOI: 10.1007/s00253-008-1628-6
Tonin, F., Melis, R., Cordes, A., Sanchez-Amat, A., Pollegioni, L., and Rosini, E. (2016). “Comparison of different microbial laccases as tools for industrial uses,” New Biotechnology 33(3), 387-398. DOI: 10.1016/j.nbt.2016.01.007
Vilar, J. M., Guet, C. C., and Leibler, S. (2003). “Modeling network dynamics the lac operon, a case study,” The Journal of Cell Biology 161(3), 471-476. DOI: 10.1083/jcb.200301125
Wagner, S., Klepsch, M. M., Schlegel, S., Appel, A., Draheim, R., Tarry, M., Högbom, M., Van Wijk, K. J., Slotboom, D. J., and De Gier, J. W. (2008). “Tuning Escherichia coli for membrane protein overexpression,” Proceedings of the National Academy of Sciences U.S.A. 105(38), 14371-14376. DOI: 10.1073/pnas.0804090105.
Wang, H., Wang, F., Wang, W., Yao, X., Wei, D., Cheng, H., and Deng, Z. (2014). “Improving the expression of recombinant proteins in E. coli BL21 (DE3) under acetate stress: an alkaline pH shift approach,” PLOS ONE 9(11), e112777. DOI: 10.1371/journal.pone.0112777
Wang, Y., Bouchabke-Coussa, O., Lebris, P., Antelme, S., Soulhat, C., Gineau, E., Dalmais, M., Bendahmane, A., Morin, H., Mouille, G., Legèe, F., Cézard, L., Lapierre, C., and Sibout, R. (2015). “LACCASE5 is required for lignification of the Brachypodium distachyon Culm,” Plant Physiology 168(1), 192-204. DOI: 10.1104/pp.114.255489
Wurm, D. J., Veiter, L., Ulonska, S., Eggenreich, B., Herwig, C., and Spadiut, O. (2016). “The E. coli pET expression system revisited—Mechanistic correlation between glucose and lactose uptake,” Applied Microbiology and Biotechnology 1-9. DOI: 10.1007/s00253-016-7620-7.
Yang, J., Wang, G., Ng, T. B., Lin, J., and Ye, X. (2015). “Laccase production and differential transcription of laccase genes in Cerrena sp. in response to metal ions, aromatic compounds, and nutrients,” Frontiers in Microbiology 6. DOI: 10.3389/fmicb.2015.01558
Yao, Y., Sakamoto, T., Honda, Y., Kagotani, Y., Izumitsu, K., Suzuki, K., and Irie, T. (2013). “The white-rot fungus Pleurotus ostreatus transformant overproduced intracellular cAMP and laccase,” Bioscience, Biotechnology and Biochemistry 77(11), 2309-2311. DOI: 10.1271/bbb.130470
Ye, Y. X., Pan, P. L., Kang, D., Lu, J. B., and Zhang, C. X. (2015). “The multicopper oxidase gene family in the brown planthopper, Nilaparvata lugens,” Insect Biochemistry and Molecular Biology 63, 124-132. DOI: 10.1016/j.ibmb.2015.06.010.
Zhang, Y. B., Jiang, M. L., Hu, X. J., Zhang, G. M., and Ma, L. X. (2005). “Expression of a laccase gene from Pleurotus ostreatus in Pichia pastoris and characterization of the recombinant enzyme,” Wei sheng wu xue bao= Acta microbiologica Sinica 45(4), 625-629.
Article submitted: November 24, 2016; Peer review completed: January 28, 2017; Revised version received and accepted: March 8, 2017; Published: March 13, 2017.
DOI: 10.15376/biores.12.2.3211-3221
