Abstract
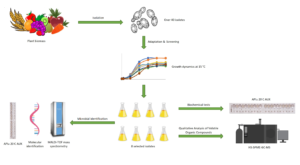
More than 40 yeast strains were isolated from various types of plant biomass and then evaluated for potential applications in biotechnological processes conducted at low temperature. Adaptation to low temperature was tested by passaging the isolates at decreasing temperatures, from 30 to 15 °C. Only the strains that were able to adapt to the final temperature and reached the stationary growth phase relatively quickly were submitted to further experimentation. These included eight environmental yeast isolates from four types of materials of plant origin: wheat, rye, and cucumber, containing glucose, fructose, sucrose, and starch; yeast-fermentable sugars; red beetroot, containing large amounts of glucose and fructose; and fruits (grapes and apples) containing glucose, fructose, and sucrose. The strains were identified and then subjected to a series of experiments to assess their suitability for use in low-temperature biotechnological industrial processes incorporating microbial biomass. The growth dynamics and assimilation profiles of the yeast strains were investigated, as well as their ability to produce volatile compounds.
Download PDF
Full Article
Plant Biomass as a Source of Low-Temperature Yeasts
Wiktoria Liszkowska,a,* Ilona Motyl,a Katarzyna Pielech-Przybylska,b Justyna Szulc,a Marcin Sypka,c Piotr Dziugan,a and Joanna Berłowska a,*
More than 40 yeast strains were isolated from various types of plant biomass and then evaluated for potential applications in biotechnological processes conducted at low temperature. Adaptation to low temperature was tested by passaging the isolates at decreasing temperatures, from 30 to 15 °C. Only the strains that were able to adapt to the final temperature and reached the stationary growth phase relatively quickly were submitted to further experimentation. These included eight environmental yeast isolates from four types of materials of plant origin: wheat, rye, and cucumber, containing glucose, fructose, sucrose, and starch; yeast-fermentable sugars; red beetroot, containing large amounts of glucose and fructose; and fruits (grapes and apples) containing glucose, fructose, and sucrose. The strains were identified and then subjected to a series of experiments to assess their suitability for use in low-temperature biotechnological industrial processes incorporating microbial biomass. The growth dynamics and assimilation profiles of the yeast strains were investigated, as well as their ability to produce volatile compounds.
DOI: 10.15376/biores.18.1.599-612
Keywords: Yeast; Plant biomass; Volatile organic compounds; Metabolic profile; Fermentation; Low temperature
Contact information: a: Department of Environmental Biotechnology, Lodz University of Technology,
90-924 Lodz, Poland; b: Institute of Fermentation Technology and Microbiology, Lodz University of Technology, 90-530 Lodz, Poland; c: Institute of Molecular and Industrial Biotechnology, 90-573 Lodz, Poland; *Corresponding authors: joanna.berlowska@p.lodz.pl; wiktoria.liszkowska@dokt.p.lodz.pl
GRAPHICAL ABSTRACT

INTRODUCTION
The term biomass was first introduced by the U.S. Congress in the Powerplant and Industrial Fuel Use Act of 1978, as a type of alternate fuel. Later, the term was defined more precisely as “any organic matter which is available on a renewable basis, including agricultural crops and agricultural wastes and residues, wood and wood wastes and residues, animal wastes, municipal wastes, and aquatic plants.” Although biomass is often seen primarily as a source of heat and electricity (Bracmort 2013), there is increasing interest in using this organic matter from wastes as a potential source of organic compounds (Maurya et al. 2015). Biomass can be classified based on the various scientific categories. In general, biomass is derived from industrial processes, and can be classified as:
- Agricultural biomass (wheat, beetroot, sugar cane, maize, etc.), animal biomass (such as fats), and energy crops
- Agricultural and livestock co-products and residues (straw, pulp, manure, etc.)
- Fisheries and aquaculture biomass – animal products and waste from the water environment, algae, and microalgae
- Forest wood, energy tree plantations
- Waste from the wood industry, woods, and forestry
- Waste from the food industry, housing, and urban communities (Shankar et al. 2011; Sourisse 2017)
Another classification system focuses on naturally occurring biomass of plant origin (not waste derived from industry). This short, more detailed classification is used to characterize the types of biomass used in the production of chemicals, fuels, and power:
- Grains – a source of carbohydrates, good material for first-generation biofuel, fat, and protein production
- Lignocellulosic materials – a source of cellulose, hemicellulose, and lignin, good material for second-generation biofuel and paper production
- Seeds – used for first-generation biofuel production
- Algae – an alternative source for biofuel production (Muhammad et al. 2013; Martín 2016)
A summary of the biomass categories is presented in Fig. 1.
Fig. 1. Sources of biomass
Biomass is a potential source of many chemical compounds. It can be used to produce various groups of components, such as amino acids, proteins, microelements, and macroelements, as well as vitamins (Guimarães et al. 2007; Scott et al. 2007). Natural biomass can also be used as an environment for the biosynthesis of volatile organic compounds, which are commercially important (Schwab et al. 2008). Carbohydrates are key components for the microbial biotransformation of agricultural biomass such as crops in the food industry (Guimarães 2012).
Microorganisms of various origins are used in biotechnological processes related to biomass processing. Microbes, including yeasts and bacteria, are able to use saccharides as a carbon source for growth, proliferation, and production of enzymes. The most important group of enzymes for yeasts are glycoside hydrolases, especially invertase, amylase, and maltase (Guimarães et al. 2007; Reis et al. 2014). Other enzymes forming secondary metabolites (such as transferases and esterases) are also very important for the food industry, due to their production of various aroma compounds.
Environmental isolates are very resistant to factors affecting cultivation. They can adapt faster to new conditions, because in their natural habitats they are exposed to environmental changes (Bleuven and Landry 2016; Kumar et al. 2021). An example is the vineyard environment, in which yeasts meet new challenges and continuously adapt to the changing conditions induced by human intervention. Isolates from such environments can be involved in wide range of processes. A good example of those yeasts are non-Saccharomyces yeasts, such as Hanseniaspora uvarum or Metschnikowia pulcherrima, which are wild-type species able to initiate glucose utilization at the early stage of wine making. They are desired to produce volatile aroma compounds that influence the sensory profile of wine. Another process that relies on the spontaneous fermentation by wild yeasts is traditional beer production, in particular Belgian Lambic beers. In the fermentation process, wild yeasts belonging to the genus Dekkera spontaneously ferments wort (Steensels and Verstrepen 2014).
In this study, more than 40 yeast strains were isolated from various sources of plant biomass. Eight of the isolates were identified and then evaluated for potential applications in biotechnological processes at low temperatures. The growth dynamics and assimilation profiles of the yeast strains were investigated, as well as their ability to produce volatile compounds.
EXPERIMENTAL
Eight yeast strains were isolated from natural plant sources: pickled beetroot, pickled cucumber, wheat flour, rye flour, grapes, and natural apple juice. The strains were cultured in 10 mL of wort broth (Merck, Darmstadt, Germany) in test tubes at 15 °C for 5 days.
Table 1. Yeast Isolates Used in Experimental Part
Microbial Identification Using MALDI-TOF Mass Spectrometry
For microbial identification, an AXIMA-iD Plus Confidence MALDI-TOF MS System (Kratos Analytical Ltd and Shimadzu Corporation, Kyoto, Japan) was used with SARAMIS Premium software (Spectral Archive and Microbial Identification System, bioMérieux, France). Yeast isolates were grown at 30 °C for 16 h on solid wort broth plates. The isolates were analyzed using the direct smear plus formic acid method provided by the producer. Microbial colonies were spread on dedicated analytical metal plates with a sterile 1 µL inoculation loop. Then, 0.5 µL of 25% formic acid was spotted onto the cells, mixed well, and left until almost dry. Subsequently, 1 μL of saturated α-cyano-4-hydroxycinnamic acid (α-CHCA) solution in an acetonitrile:ethanol:water (1:1:1 v/v) mixture containing 3% trifluoroacetic acid (TFA) was added to the disrupted microbial cells, mixed well again, and air-dried at room temperature.
The mass spectra were acquired and processed using Launchpad 2.9 software (Kratos Analytical Ltd. and Shimadzu Corporation, Kyoto, Japan) in the SARAMIS linear positive mode with a laser frequency of 50 Hz and a mass-to-charge ratio (m/z) ranging from 2,000 to 20,000 Da (laser power, 90; profiles, 200 per sample; 5 shots accumulated per profile) for each mass spectrum. As recommended by the manufacturer, E. coli DH5α (TAKARA BIO INC., Shiga, Japan) cells were used as a calibrator for the AXIMA-iD Plus Confidence MALDI-TOF MS System and as an internal control for the identification process.
Molecular Identification of Isolated Yeasts
Single yeast colonies, cultured on wort broth medium (Merck, Darmstadt, Germany) for 24 h at 30 °C, were used in the colony PCR procedure. Amplification of the internal transcribed spacers (ITS1 and ITS2 regions), including 5.8S rRNA gene, and the D1/D2 domains of the large subunit (LSU) rRNA gene region was performed by cycle sequencing using the following forward and reverse primers, respectively: ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) for ITS regions; and NL1FWD (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL4REV (5′-GGT CCG TGT TTC AAG ACG G-3′) for the LSU D1/D2 region. The GoTaq® G2 Hot Start Polymerase Kit (Promega, USA) was used for PCR. After the reaction, the product was cleaned with the commercially available GeneMATRIX Basic DNA Purification Kit (EURx, Poland). The amplified fragments were sequenced by Genomed S.A. (Warsaw, Poland) using previously described forward and reverse primers. The sequences were aligned, analyzed, and corrected, and compared with sequences from the GenBank database.
Biochemical Tests
Biochemical tests were conducted using a standardized API® 20 C AUX commercial test (bioMérieux, France), according to the manufacturer’s instructions. The API 20 C AUX strip contains 19 couples with dehydrated substrates as carbon sources for yeasts. If the given sugar is assimilated by the given yeast strain, turbidity occurs in the microtubes. The strips were incubated at 15 °C and 30 °C.
Yeasts Growth Dynamics
The growth dynamics of the environmental yeast isolates were studied by determining the optical density of yeast suspension (Densitometer DEN-1B, Grant Instruments (Cambridge) Ltd, Shepreth, UK) and the number of CFU/mL. Small amounts of the biomass from each yeast isolate were transferred to a wort broth medium. The optical density of each culture was measured. In the next step, serial dilutions in saline were prepared and Petri dishes were inoculated. The cultures were incubated at 15 °C for 5 days, after which the colonies were counted. This procedure was repeated every 24 h until the optical density value leveled out.
Qualitative Analysis of Volatile Organic Compounds Using the HS-SPME-GC-MS Technique
Volatile organic compounds (VOCs) were identified by the gas chromatography method with mass spectrometry (GC-MS), using solid phase microextraction (SPME) of VOCs from 5-day cultures of yeast in wort broth medium, incubated at 15 °C. For this purpose, 5 mL samples in tightly closed 20-mL glass vials were thermostated in a block placed on a magnetic stirrer with a heating function. The process was carried out at 50 °C, with a constant stirring speed of 400 rpm. After this time, CAR/PDMS/DVB fiber (2 cm, 50/30 µm) was placed in the vial and VOCs were extracted from the headspace for 15 min at 50 °C. Subsequently, the fiber was transferred to a gas chromatograph injector.
Desorption of volatile analytes was carried out at 250 °C for 5 min. The injector had the function of dividing the stream of carrier gas (split 2:1). The separation of compounds was carried out on a capillary column (DB1-MS, Agilent, Santa Clara, CA, USA) with dimensions of 60 m (length) × 250 μm (internal diameter) × 0.25 μm (thickness of the stationary phase), using a programmed temperature increase of 30 °C for 6 min, increasing 5 °C/min up to 230 °C and remaining at 230 °C for 2 min. The flow of carrier gas (helium) through the column was 1.1 mL/min. The temperatures of the GC injector, transfer line to MS, ion source, and quadrupole were 250, 250, 230, and 150 °C, respectively. During analysis, the mass spectrometer was operating in the full scan mode. Identification of volatile compounds was performed by comparison of obtained spectra with the reference mass spectra from NIST/EPA/NIH mass spectra library (2012; Version 2.0 g) and with the mass spectra of the reference substances. Based on SPME-GC-MS analysis results, the mean relative peak areas of identified compounds was calculated (expressed as % of total peak areas). Data processing was conducted with Mass Hunter Workstation Software (Agilent, Santa Clara, CA, USA).
SPME-GC-MS analyses were performed in triplicate. Since the results of GC-MS analysis were non-normally distributed (Shapiro-Wilk’s test), the non-parametric Kruskal–Wallis test, with Dunn’s multiple comparisons test, was used to examine differences (p ≤0.05). Statistical analysis were performed using XLSTAT software (Addinsoft, version 2022.2.1, USA).
RESULTS AND DISCUSSION
There is a growing body of scientific literature on the adaptation of yeasts to low temperatures. Environmental yeast isolates are able to adapt easily to changing environmental conditions. This makes them valuable for use in biotechnological processes carried out at low temperatures. In the present study, the tested yeast strains were found to grow well at 15 ℃, making them suitable for various processes in the food industry. For example, yeasts are commonly used at temperatures of up to 15 °C to produce bottom-fermented lager beers and cider (Vidgren et al. 2010). The sensory profiles of low-temperature fermented alcohols are enriched with large amounts of esters and other valuable volatile organic compounds (Gamero et al. 2014). This well-known process could benefit from broadening the operational spectrum of low-temperature yeasts. Other biotechnological operations also have advantages when conducted at low temperatures. Although there is a lower increase in biomass than at temperatures of around 30 ℃, from the microbiological point of view low-temperature processes provide more efficient substrate-product conversion (limiting undesired by-products), reduce the degradation of thermolabile products, and inhibit the growth of undesired microflora (Liszkowska and Berlowska 2021).
In this study, eight yeasts were chosen out of 40 isolates for their ability to reach stationary phase in a relatively short time. The eight yeasts were identified based on their assimilation profiles using the API® 20 C AUX test, the MALDI-TOF mass spectrometry method and molecular identification. API tests are commonly used to characterize assimilated sugars and for initial identification of microorganisms based on the assimilation pattern. The analysis is rather inexpensive, but it requires several days (Spencer et al. 2011). The results are reliable, but they may be affected by the diversity of natural existing microflora and environmental conditions. The assimilation profile was examined at the recommended temperature. Initial identification of the yeast strains was performed by biochemical tests. The second identification method was MALDI-TOF mass spectrometry. Metabolomic identification has a higher probability of success than conventional methods. Identification is based on the proteome, a unique set of proteins in the analyzed microorganism. This proteome is like a fingerprint of the cell (Singhal et al. 2015). This method is accurate, very sensitive, and can be performed in 5 to 10 min. By comparison, standard biochemical methods take 5 to 48 h. Metabolomic identification can be performed using a single colony. As a reference, the most reliable method, molecular identification was carried out.
Table 2. Identification of Yeast Isolates
Table 2 gives the names of the identified strains and the confidence score values (compared to data deposited in the database). Confidence scores in MALDI-TOF mass spectrometry are considered excellent if they are higher than 99.9%; good scores are between 85% and 99.8%; scores of 70 to 84.9% are considered sufficient.
Molecular identification was used as a reference method. Seven of the eight isolates were identified with great confidence. Yeast strain 10 was not identified. The problem with sequencing may be related to the source of isolate – environmental plant biomass. It may contain many nucleotide polymorphisms that make the identification impossible.
Identified yeast isolates were compared to the following sequences deposited in GenBank:
- Kazachstania barnetti – AY046173.1
- Saccharomyces cerevisiae – KT764941.1 (isolate 2) and MT322849.1 (isolate 13)
- Pichia fermentans – KY104537.1
- Hanseniaspora uvarum – KY103573.1 (isolate 9)
- Wickerhamomyces anomalus – MZ351763.1 (isolate 11) and JX188245.1 (isolate 12).
In MALDI-TOF mass spectrometry method, seven analyzed isolates were positively matched and compared to the reference test. In the proteomic method, the scores for samples 2, 11, 12, and 13 were perfect matches, whereas samples 8 and 9 were identified with good confidence. Yeast strain 10 was not identified with sufficient confidence. However, based on the isolation source (grapes) and metabolic features, it can be assumed to have been identified correctly.
Comparing both methods, the obtained results were consistent. Six of the eight isolates have been identified accurately. The differences in taxonomic names are probably due to the limitation of the mass spectra database (Launchpad 2.9 software), in which data from MALDI-TOF mass spectrometry experiment are processed. However, it can be checked that identified strains are considered to be one and the same – Candida lambica is a heterotypic synonym of Pichia fermentans and Candida pelliculosa is a heterotypic synonym of Wickerhamomyces anomalus.
For basic identification, API® 20 C AUX biochemical tests were used (Table 2). This method provided slightly different results to those from metabolomic and molecular identification. Only three of the eight yeast isolates were classified as being the same genus as analyzed by molecular identification and MALDI-TOF mass spectrometry with high confidence: samples 2, 9, 13. The results for isolate 8 were completely different. Nonetheless, the biochemical tests provided important information for classifying the environmental isolates and identifying the carbon sources they assimilated. Assimilation tests are valuable for indicating the metabolic abilities of yeasts and may indicate potential applications (raw materials for culturing). Their use for initial identification can be treated as an added value.
According to the literature, seven of the eight isolated strains have been confirmed to occur naturally. Pichia fermentans is found on vegetables, such as cucumbers (Strausbaugh and Gillen 2008; Douglass et al. 2018). Hanseniaspora uvarum occurs on grapes (Albertin et al. 2016). Wickerhamomyces anomalus is found in wheat flour (Li et al. 2021). Saccharomyces cerevisiae occurs in rye flour and apple (Rosenquist and Hansen 2000; Suárez Valles et al. 2005). Yeast isolate 1 (Kazachstania barnetti) has not been found yet on beetroots, so it seems to be a novel strain occurring in this type of plant biomass.
Many factors can influence yeast growth, such as oxygen conditions, the availability of nitrogen and carbon source, pH, water activity, and temperature. In this study, temperature was an evaluation factor during screening. We examined how quickly the environmental yeast isolates adapted to below optimum temperatures.
The growth dynamics of the isolated yeasts at 15 °C were assessed using two methods: by measuring optical density of yeast suspensions and by counting the number of CFU/mL. The results are presented in Figs. 2 and 3.
The investigated yeast isolates reached the stationary phase between 72 and 96 h of culturing. K. barnetti and S. cerevisiae 1 showed the highest growth of biomass (above 108 CFU/mL). The lowest growth was observed for P. fermentans.
Fig. 2. Growth dynamics at 15 °C – optical density measurement
Fig. 3. Growth dynamics at 15 °C – CFU/mL
The API® 20 C AUX test is a good tool for evaluating the assimilation profiles of yeasts. It was therefore used to determine the ability of the enzymes to utilize given carbohydrates as a carbon source at 30 and 15 °C. The results are presented in Tables 3 and 4. Sugars that were not assimilated by any strain are not included in the tables.
Table 3. Assimilation Profiles of the Yeast Isolates at 30 °C
Table 4. Assimilation Profiles of Yeast Isolates at 15 °C
The assimilation profile at 15 ℃ was lower than that obtained at 30 ℃. As expected, all the tested yeast isolates were able to utilize glucose (GLU) as a carbon source. The strains isolated from similar types of biomass showed similar biochemical profiles. H. uvarum 1 and 2 from grapes were able to assimilate, apart from glucose, calcium 2-keto gluconate (2KG) and cellobiose (CEL). W. anomalus 1 and 2 showed almost the same profiles: the ability to utilize glucose, glycerol (GLY), methyl-α-D-glucopyranoside (MDG), maltose (MAL), and sucrose (SAC). Additionally, W. anomalus 1 was able to use D-melezitose (MLZ). The yeasts from pickled vegetables, K. barnetti and P. fermentans, presented comparable profiles (glucose and N-acetyl-glucosamine). However, K. barnetti from pickled red beetroot was also able to assimilate D-xylose (XYL). Interestingly, the low-temperature conditions did not induce assimilation of trehalose.
Table 5. Results of HS-SPME-GC-MS Qualitative Analysis of Volatile Organic Compounds Formed at 15 °C
Depending on their content and concentration, volatile organic compounds (VOCs) contribute positively or negatively to the sensory characteristics of food products, affecting food quality. Their aroma profile depends on temperature and culture conditions (Peng et al. 2015; Carrau et al. 2017). We used the HS-SPME-GC-MS technique to assess the formation of VOCs by the selected yeast isolates. This method provides a fast and reliable way to qualitatively identify large numbers of VOCs from relatively small samples (Diez-Simon et al. 2020). Wort broth of a known composition, provided by the manufacturer, was used as a medium. Samples for analysis were collected after 5 days of culturing at
15 °C.
Qualitative analysis showed that a broad spectrum of volatile compounds were synthesized during growth of the yeasts. At least 70 different VOCs were present in the samples, 20 of which constituted at 1% of the total amount of identified compounds. The results are presented in Table 5. Compounds that constituted less than 1% of the chromatogram are not included in the table.
Qualitative analysis revealed that almost all the compounds belonged to the ester group. Most of these esters occurred in the medium with H. uvarum 1, W. anomalus 1 and W. anomalus 2. H. uvarum is an isolate from grapes, and W. anomalus 1 and 2 are isolates from wheat flour. The esters comprised around 60% of all the compounds in the three samples. Esters such as ethyl hexanoate, ethyl acetate, ethyl dodecanoate, and esters of butanol are typical compounds formed by yeast (Harman et al. 2021). In the cases of K. barnetti, S. cerevisiae 2 and P. fermentans, yeast isolates from pickled red beetroot, rye flour, and pickled cucumber, typical alcohols such as butyl alcohols comprised two or three times more of the compounds than esters.
Based on these results, the yeast isolates cultured under similar conditions but at temperatures below optimum (25 to 35 °C) had slightly different biochemical abilities and growth dynamics (Siew et al. 2007). The yeasts were capable of successful adaptation to low temperature while maintaining good metabolic activity. The biosynthesis of aroma compounds and growth dynamics were slightly prolonged. However, the results including the amount of biomass are satisfactory.
Environmental yeast isolates from various environments are considered to be good biological material with potential applications in various industries. They can acclimate easily to new environmental conditions. Wild yeasts can also have a beneficial impact on aroma profile (Di Paola et al. 2020). For instance, Pichia anomala and Pichia fermentans yeasts have been investigated to enrich wine in volatile esters and higher alcohols (Steensels and Verstrepen 2014). There are ongoing studies of isolation and identification of wild type yeasts from different sources. There is a growing focus on such genera and their influence on fermentation process, for instance Saccharomyces, Pichia, Candida, and Kazachstania genera affecting on the products made by traditional methods (Beyene et al. 2020). This leads to the conclusion that biomass is a good source of wild-type microorganisms, which could have beneficial applications, in particular in fermented food and beverages production.
CONCLUSIONS
- Yeast isolates possessed satisfactory metabolic activity at temperatures below optimum. The stationary growth phase was reached within 72 to 96 h, enabling further low-temperature biotechnological processes.
- The sensory profile at 15 °C was characterized by a high content of esters, which are desirable in fermentation processes, especially in the food and cosmetics industries.
- Biosynthesis of volatile organic compounds conducted by tested yeast isolates must be analyzed quantitatively and optimized to intensify the formation of desired compounds.
ACKNOWLEDGMENTS
This research was completed while the first and fifth authors were the doctoral candidates in the Interdisciplinary Doctoral School at Lodz University of Technology, Poland.
REFERENCES CITED
Albertin, W., Setati, M. E., Miot-Sertier, C., Mostert, T. T., Colonna-Ceccaldi, B., Coulon, J., Girard, P., Moine, V., Pillet, M., Salin, F., Bely, M., Divol, B., and Masneuf-Pomarede, I. (2016). “Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering,” Frontiers in Microbiology 6(JAN), article 1569. DOI: 10.3389/FMICB.2015.01569/BIBTEX
Beyene, E., Tefera, A. T., Muleta, D., Fantahun, S. K., and Wessel, G. M. (2020). “Molecular identification and performance evaluation of wild yeasts from different Ethiopian fermented products” Journal of Food Science and Technology 57, 3436-3444. DOI: 10.1007/s13197-020-04377-7
Bleuven, C., and Landry, C. R. (2016). “Molecular and cellular bases of adaptation to a changing environment in microorganisms,” Proceedings of the Royal Society B: Biological Sciences 283(1841). DOI: 10.1098/RSPB.2016.1458
Bracmort, K. (2013). “Biomass: Comparison of Definitions in Legislation,” published on Nov. 22, 2012 and updated on June 27, 2019.
Carrau, F., Boido, E., and Dellacassa, E. (2017). “Yeast diversity and flavor compounds,” Fungal Metabolites, Springer, Cham, 569-597. DOI: 10.1007/978-3-319-25001-4_32
Diez-Simon, C., Ammerlaan, B., van den Berg, M., van Duynhoven, J., Jacobs, D., Mumm, R., and Hall, R. D. (2020). “Comparison of volatile trapping techniques for the comprehensive analysis of food flavourings by gas chromatography-mass spectrometry,” Journal of Chromatography A 1624, 461191. DOI: 10.1016/J.CHROMA.2020.461191
Di Paola, M., Meriggi, N., and Cavalieri, D. (2020). “Applications of wild isolates of Saccharomyces yeast for industrial fermentation: The gut of social insects as niche for yeast hybrids’ production,” Frontiers in Microbiology 11, 2674. DOI: 10.3389/FMICB.2020.578425/BIBTEX
Douglass, A. P., Offei, B., Braun-Galleani, S., Coughlan, A. Y., Martos, A. A. R., Ortiz-Merino, R. A., Byrne, K. P., and Wolfe, K. H. (2018). “Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names,” PLOS Pathogens 14(7), article e1007138. DOI: 10.1371/JOURNAL.PPAT.1007138
Gamero, A., Ferreira, V., Pretorius, I. S., and Querol, A. (2014). “Wine, beer and cider: Unravelling the aroma profile,” Molecular Mechanisms in Yeast Carbon Metabolism, Springer-Verlag Berlin Heidelberg, 261-297. DOI: 10.1007/978-3-642-55013-3_10/COVER
Guimarães, L. H. S. (2012). “Carbohydrates from biomass: Sources and transformation by microbial enzymes,” Carbohydrates – Comprehensive Studies on Glycobiology and Glycotechnology, IntechOpen. DOI: 10.5772/51576
Guimarães, L. H. S., Terenzi, H. F., Polizeli, M. de L. T. de M., and Jorge, J. A. (2007). “Production and characterization of a thermostable extracellular β-d-fructofuranosidase produced by Aspergillus ochraceus with agroindustrial residues as carbon sources,” Enzyme and Microbial Technology 42(1), 52-57. DOI: 10.1016/J.ENZMICTEC.2007.07.021
Kumar, V., Sarma, V. V., Thambugala, K. M., Huang, J. J., Li, X. Y., and Hao, G. F. (2021). “Ecology and evolution of marine fungi with their adaptation to climate change,” Frontiers in Microbiology 12, 2527. DOI: 10.3389/FMICB.2021.719000/BIBTEX
Li, Z., Zhou, M., Cui, M., Wang, Y., and Li, H. (2021). “Improvement of whole wheat dough fermentation for steamed bread making using selected phytate-degrading Wickerhamomyces anomalus P4,” Journal of Cereal Science 100, 103261. DOI: 10.1016/j.jcs.2021.103261
Liszkowska, W., and Berlowska, J. (2021). “Yeast fermentation at low temperatures: Adaptation to changing environmental conditions and formation of volatile compounds,” Molecules 26(4), 1035. DOI: 10.3390/MOLECULES26041035
Martín, M. M. (2016). “Biomass,” Industrial Chemical Process Analysis and Design, Elsevier, 405-447. DOI: 10.1016/B978-0-08-101093-8.00022-7
Maurya, D. P., Singla, A., and Negi, S. (2015). “An overview of key pretreatment processes for biological conversion of lignocellulosic biomass to bioethanol,” 3 Biotech 5(5), 597-609. DOI: 10.1007/S13205-015-0279-4/TABLES/1
Muhammad, H., Iqbal, N., Kyazze, G., and Keshavarz, T. (2013). “Biotech applications of biomass,” BioResources 8(2), 3157-3176
Peng, B., Li, F., Cui, L., and Guo, Y. (2015). “Effects of fermentation temperature on key aroma compounds and sensory properties of apple wine,” Journal of Food Science 80(12), S2937-S2943. DOI: 10.1111/1750-3841.13111
Reis, B. A. D., Kosińska-Cagnazzo, A., Schmitt, R., and Andlauer, W. (2014). “Fermentation of plant material – Effect on sugar content and stability of bioactive compounds,” Polish Journal of Food and Nutrition Sciences 64(4), 235-241. DOI: 10.2478/PJFNS-2013-0017
Rosenquist, H., and Hansen, Å. (2000). “The microbial stability of two bakery sourdoughs made from conventionally and organically grown rye,” Food Microbiology 17(3), 241-250. DOI: 10.1006/FMIC.1999.0313
Schwab, W., Davidovich-Rikanati, R., and Lewinsohn, E. (2008). “Biosynthesis of plant-derived flavor compounds.” DOI: 10.1111/j.1365-313X.2008.03446.x
Scott, E., Peter, F., and Sanders, J. (2007). “Biomass in the manufacture of industrial products—the use of proteins and amino acids,” Applied Microbiology and Biotechnology 75(4), 751. DOI: 10.1007/S00253-007-0932-X
Shankar, J., Shahab, T., Christopher, S., Wright, T., Boardman, R. D., and Yancey, N. A. (2011). “A review on biomass classification and composition, co-firing issues and pretreatment methods,” in: 2011 ASABE Annual International Meeting.
Siew, L. T., Daran-Lapujade, P., Walsh, M. C., Pronk, J. T., and Daran, J. M. (2007). “Acclimation of Saccharomyces cerevisiae to low temperature: A chemostat-based transcriptome analysis,” Molecular Biology of the Cell 18(12), 5100. DOI: 10.1091/MBC.E07-02-0131
Singhal, N., Kumar, M., Kanaujia, P. K., and Virdi, J. S. (2015). “MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis,” Frontiers in Microbiology 6(AUG). DOI: 10.3389/FMICB.2015.00791
Sourisse, C. (2017). Les ressources du futur issues du monde végétal – Vers un nouveau système de production de nourriture, d’énergie, et de matériaux, Covabis, Saint-Ismier.
Spencer, J., Rawling, S., Stratford, M., Steels, H., Novodvorska, M., Archer, D. B., and Chandra, S. (2011). “Yeast identification: Reassessment of assimilation tests as sole universal identifiers,” Letters in Applied Microbiology 53(5), 503-508. DOI: 10.1111/J.1472-765X.2011.03130.X
Steensels, J., and Verstrepen, K. J. (2014). “Taming wild yeast: Potential of conventional and nonconventional yeasts in industrial fermentations,” Annual Review of Microbiology 68, 61-80. DOI: 10.1146/annurev-micro-091213-113025
Strausbaugh, C. A., and Gillen, A. M. (2008). “Bacteria and yeast associated with sugar beet root rot at harvest in the Intermountain West,” Plant Disease 92(3), 357-363. DOI: 10.1094/PDIS-92-3-0357
Suárez Valles, B., Pando Bedriñana, R., Fernández Tascón, N., González Garcia, A., and Rodríguez Madrera, R. (2005). “Analytical differentiation of cider inoculated with yeast (Saccharomyces cerevisiae) isolated from Asturian (Spain) apple juice,” LWT – Food Science and Technology 38(5), 455-461. DOI: 10.1016/J.LWT.2004.07.008
Vidgren, V., Multanen, J. P., Ruohonen, L., and Londesborough, J. (2010). “The temperature dependence of maltose transport in ale and lager strains of brewer’s yeast,” FEMS Yeast Research 10(4), 402-411. DOI: 10.1111/J.1567-1364.2010.00627.X
Article submitted: August 26, 2022; Peer review completed: September 18, 2022; Revised version received and accepted: November 13, 2022; Published: November 21, 2022.
DOI: 10.15376/biores.18.1.599-612
