Abstract
Cellulolytic enzymes from fungi are complex compound enzyme structures that generally include three kinds of hydrolytic enzymes, which are called endo-β-1,4-glucanases, exo-β-1,4-glucanases, and β-1,4-glucosidases (β-glucosidases). The fungus Mucor ardhlaengiktus isolated from field rice straw produced a β-glucosidase (MaBgl). Maximal MaBgl production occurred when M. ardhlaengiktus was incubated for 6 days in fermentation liquor at 30 °C and an initial pH of 6. Purified MaBgl was obtained from M. ardhlaengiktus fermentation broth by ammonium sulfate fractional precipitation and DEAE-Sepharose FF ion exchange chromatography. The molecular weight of MaBgl as determined by SDS-PAGE electrophoresis was approximately 72 kDa. The kinetic parameters, Michaelis constant (Km) and maximum velocity (Vmax), of MaBgl were 78.2 μmol/L and 28.5 μmol/(L·min), respectively. Assays of MaBgl produced by M. ardhlaengiktus RSC1 under different conditions were investigated by the 3,5-dinitrosalicylic acid (DNS) assay for glucose. The pH and temperature optima for catalytic activity of MaBgl were pH 4.8 and 50 °C, respectively. MaBgl exhibited good thermal stability in the range of 20 to 30 °C, but the thermal stability of MaBgl decreased rapidly over 60 °C. MaBgl had better pH stability between pH 4.6 and 5.0, and the stability of MaBgl decreased when the pH value was lower or higher than this range.
Download PDF
Full Article
Characteristics and Kinetic Analysis of β-glucosidase (MaBgl) from Mucor ardhlaengiktus RSC1
Zhihai Yang,a, Liping Wu,b, Mingjia Fu,a,* Qin Li,a and Dexiao Ye a
Cellulolytic enzymes from fungi are complex compound enzyme structures that generally include three kinds of hydrolytic enzymes, which are called endo-β-1,4-glucanases, exo-β-1,4-glucanases, and β-1,4-glucosidases (β-glucosidases). The fungus Mucor ardhlaengiktus isolated from field rice straw produced a β-glucosidase (MaBgl). Maximal MaBgl production occurred when M. ardhlaengiktus was incubated for 6 days in fermentation liquor at 30 °C and an initial pH of 6. Purified MaBgl was obtained from M. ardhlaengiktus fermentation broth by ammonium sulfate fractional precipitation and DEAE-Sepharose FF ion exchange chromatography. The molecular weight of MaBgl as determined by SDS-PAGE electrophoresis was approximately 72 kDa. The kinetic parameters, Michaelis constant (Km) and maximum velocity (Vmax), of MaBgl were 78.2 μmol/L and 28.5 μmol/(L·min), respectively. Assays of MaBgl produced by M. ardhlaengiktus RSC1 under different conditions were investigated by the 3,5-dinitrosalicylic acid (DNS) assay for glucose. The pH and temperature optima for catalytic activity of MaBgl were pH 4.8 and 50 °C, respectively. MaBgl exhibited good thermal stability in the range of 20 to 30 °C, but the thermal stability of MaBgl decreased rapidly over 60 °C. MaBgl had better pH stability between pH 4.6 and 5.0, and the stability of MaBgl decreased when the pH value was lower or higher than this range.
Keywords: Mucor ardhlaengiktus; β-glucosidase; Kinetics of β-glucosidase
Contact information: a: College of Life Sciences, Jiangxi Normal University, Nanchang 330027, China; b: School of Life Science, Nanchang University, 330031, Nanchang, China; Equal contributor
* Corresponding author: mingjiafu@126.com
INTRODUCTION
Cellulose is the most abundant renewable carbon and energy source in the world. It can be naturally degraded to glucose and other saccharides with different grades of polymerization by cellulolytic microorganisms that produce cellulases, which break the -1,4-glycosidic bonds linking the glucose units in polysaccharides (Baldrian and Valášková 2008). Lignocellulosic biomass, the major combustible component of non-food energy crops and an abundant renewable resource, is a potential feedstock for bioethanol production (Cavka et al. 2014; Kiran et al. 2014; Nanda et al. 2014). Cellulolytic fungi and bacteria have complex cellulase systems that actively hydrolyze cellulose (Tomme et al. 1995). Fungal and bacterial cellulases play an important role in the biospheric carbon cycle and have many industrial applications (Bhat and Bhat 1997; Gaur and Tiwari 2015).
Cellulase, which is very effective in enzymatic hydrolysis because it acts on substrates synergistically, is usually secreted as part of multi-enzyme complexes that are made up of several proteins, including three types of cellulase components, namely, endoglucanases (EC 3.2.1.4, endo-β-D-1,4-glucanase), exoglucanases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21) (Bhat and Bhat 1997; Teeri 1997; Wilson and Irwin 1999; Lynd et al. 2002; Kumar et al. 2008; Zhang et al. 2013; El-Naggar et al. 2015b).
Enzyme production is the most crucial and costly step in the process of cellulose hydrolysis. The subdivisions Ascomycetes, Basidiomycetes, and Deuteromycetes contain large numbers of cellulolytic species (Carlile and Watkinson 1997). Member genera include Bulgaria, Chaetomium, and Helotium in Ascomycetes; Coriolus, Phanerochaete, Poria, Schizophyllum, and Serpula in Basidiomycetes; and Aspergillus, Cladosporium, Fusarium, Geotrichum, Myrothecium, Paecilomyces, Penicillium, and Trichoderma in Deuteromycetes (Carlile and Watkinson 1997). There are many fungal species that produce cellulase, including Aspergillus niger, Talaromyces pinophilus, Trichoderma koningii, Trichoderma viride, Fusarium solani, Penicillium pinophilum, and Monilia sp. (Wood and McCrae 1972; Berghem et al. 1975; Wood and McCrae 1977; Gong et al. 1981; Wood and McCrae 1986; Cavka et al. 2014; El-Naggar et al. 2015). Ortiz et al. (2015) identified a wild-type Trichoderma reesei strain that has a higher cellulase production than 19 other evaluated strains. Ang et al. (2013) and Sherief et al. (2010) have shown that Aspergillus fumigatus SK1 secretes cellulases (endoglucanase, exoglucanase, and β-glucosidase), xylanase, and protease.
In the subdivision Zygomycotina, only certain members of the genus Mucor have significant cellulolytic activity (Lynd et al. 2002). Strains of Mucor pusillus and Mucor miehei synthesize cellulase, including -glucosidase, which attacks native cellulose, acid-swollen cellulose, carboxymethylcellulose, and cellobiose (Somkuti et al. 1969; Somkuti 1974). Genomic analysis and analysis of the major PAGE gel bands of secretome proteins shows that Mucor circinelloides grows on carboxymethyl cellulose (CMC) and cellobiose, confirming the enzymatic activities of endoglucanases and -D-glucosidases, respectively (Wei et al. 2013). -Glucosidase from the soluble and wall-associated fractions of Mucor racemosus have identical physicochemical and kinetic properties (Borgia and Mehnert 1982). Borgia and Sypherd (1977) reported that M. racemosus -glucosidase activity was expressed during growth on all carbon sources studied except hexoses, whereas both hexoses and cyclic adenosine monophosphate severely repressed synthesis of the enzyme.
The current study describes the isolation of a fungal strain identified as Mucor ardhlaengiktus RSC1 from field rice straw. β-glucosidase from this strain was partially characterized, and the influence of various factors on β-glucosidase activity was analyzed. Also, β-glucosidase produced by M. ardhlaengiktus RSC1 was purified by ammonium sulfate precipitation, dialysis, and DEAE-cellulose ion-exchange chromatography. Finally, the kinetics of the purified β-glucosidase were investigated.
EXPERIMENTAL
Materials
The RSC1 strain was isolated from field rice straw in Yin Tan, Jiangxi province, China, and stored at Jiangxi Normal University. The strain was identified as M. ardhlaengiktus by 18S rDNA sequence analysis performed by the China Center of Industrial Culture Collection (CICC) and archived by CICC (strain no: CICC 2712). All chemicals and reagents were analytical grade and were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
A solution of 3,5-dinitrosalicylic acid (DNS) was prepared by first dissolving 18.2 g potassium sodium tartrate in 50 mL distilled water in a water bath at 80 ℃ until a clear solution was obtained, after which the following chemicals were added and stirred: 0.03 g 3,5-dinitrosalicylic acid, 2.1 g NaOH, and 0.5 g phenol. After cooling, the solution was brought to 100 mL with distilled water.
Methods
Preparation of crude MaBgl and assay of its activity (Ghose 1987)
The RSC1 strain was inoculated into 250-mL flasks containing 100 mL fermentation medium supplemented with 13 g/L CMC-Na, 0.8 g/L NH4NO3, 2 g/L KH2PO4, 1 g/L MgSO4, 1 g/L soybean powder, 1 g/L yeast extract, and 1 mL/L of polyoxyethylene (20) sorbitan monooleate (Tween-80). The inoculated medium was shaken at 180 rpm for 5 days at 28 ℃, after which it was centrifuged at 5000 rpm and 4 ℃ to remove the mycelia, leaving crude MaBgl enzyme in the supernatant.
MaBgl activity was assayed in a reaction mixture containing 0.5 mL of the enzyme solution and 2.0 mL of 1% salicin-disodium hydrogen phosphate-potassium dihydrogen phosphate buffer. The reaction mixture was incubated at 50 ℃ for 30 min, after which 2.5 mL of DNS solution was added. The mixture was boiled for 5.0 min, cooled, and 10 mL water was added and mixed. The OD540 of the reaction mixture was measured to determine reducing sugars. One enzyme unit (U) was defined as the enzymatic activity needed for the release of 1 μmol of glucose equivalents per unit volume and time of reaction (min).
Determination of the effect of M. ardhlaengiktus RSC1 incubation characteristics on MaBgl activity
To measure the effects of pH, temperature, and duration of the RSC1 incubation on subsequent MaBgl activity, the RSC1 fermentation and MaBgl assay procedures described above were altered with the following modifications:
- pH effect: 5 M NaOH and 5 M HCl were used to adjust the initial pH of the fermentation media to pH 5.0, pH 6.0, pH 7.0, or pH 8.0.
- Temperature effect: the RSC1 incubation was performed at temperatures of 26℃, 28 ℃, 30 ℃, 32 ℃, or 34℃, using the optimal incubation pH determined in experiment 1.
- Time effect: RSC1 incubation was performed for 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, or 7 days, using the optimal incubation pH determined in experiment 1 and temperature determined in experiment 2.
Characterization of crude MaBgl enzyme
The optimal pH for the activity of crude MaBgl enzyme was assayed as described above, but with 1% phosphate buffer adjusted to different pH values ranging from pH 4.49 to pH 9.18 before incubation at 50 ℃ for 30 min. The optimal temperature for MaBgl activity was determined at the optimal pH (determined as described above) after pre-incubation of the crude enzyme at 6 temperatures ranging from 30 ℃ to 80 ℃.
Crude MaBgl enzyme was incubated at 20, 30, 40, 50, 60, 70, 80, and 90℃ for 0.5 and 1 h respectively to determine the residual enzyme activity and determine the thermal stability of MaBgl. Crude MaBgl enzyme was mixed with pH 2.2, 2.6, 3.0, 3.4, 3.8, 4.2, 4.6, 5.0, 5.6, 6.2, 6.8, 7.4, or 8.0 citric acid-disodium hydrogen phosphate buffer solution in a certain proportion, placed at 25℃ for 1 h and 5 h, and then the residual enzyme activity was determined to determine the pH stability of MaBgl.
Purification of MaBgl
Preliminary purification of the crude MaBgl enzyme preparation was accomplished by ammonium sulfate fractional precipitation. Ammonium sulfate was added to the crude enzyme preparation to reach saturation levels of 20%, 40%, 60%, or 80%. The homogenate was centrifuged at 10,000 rpm and 4 ℃ for 20 min and then kept at 4 ℃ for 2 h. The sediment was dissolved in 0.1 M phosphate buffer (pH 5.4) followed by desalting with distilled water. Protein concentration and total MaBgl enzyme activity were measured to confirm the most suitable level of ammonium sulfate saturation. The enzyme solution was concentrated using PEG20000.
The partially purified enzyme solution was adsorbed on a DEAE-Sepharose FF column equilibrated with 0.1 M phosphate buffer (pH 7.0) and eluted by 200 mL 0.1 M potassium phosphate buffer (pH 6.0) with a linear gradient of 0 M to 0.25 M NaCl. Fractions containing MaBgl activity were collected and stored at 4℃. Protein purity and molecular mass were assessed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentration was determined by the Bradford method (Bradford 1976) using bovine serum albumin as the standard.
Determination of kinetic parameters
The Km and Vmax values were determined by a Lineweaver-Burk plot, according to a transformation of the Michaelis-Menten equation, using 1/15 M phosphate buffer (pH 4.92) to prepare a range of concentrations of salicin as a substrate (Lineweaver and Burk 1934). A mixture of 2.0 mL substrate solution and 0.5 mL enzyme solution was incubated at 50 ℃ for 0 min, 5 min, 10 min, 15 min, 20 min, 25 min, or 30 min, after which the rest of the MaBgl activity assay was performed as described above. The absorbance of the sample tube was translated into glucose production using a glucose standard curve, then glucose production and time were used to calculate V0. The double-reciprocal Lineweaver-Burk plotting method was used to obtain Km and Vmax.
RESULTS AND DISCUSSION
The Effects of M. ardhlaengiktus RSC1 Incubation Characteristics on MaBgl Activity and Production
The initial pH of the culture medium has a significant effect on the activity of β-glucosidase from several fungi (Juhász et al. 2004; Shahriarinour et al. 2011; Hmad et al. 2014; Sridevi et al. 2015; Hernández-Guzmán et al. 2016). The pH is one of the most important factors that determines the growth and morphology of fungi. MaBgl activity was assayed after M. ardhlaengiktus RSC1 had been grown for 5 days in medium with an initial pH ranging from pH 5.0 to pH 8.0. Optimal MaBgl activity was achieved when the initial pH of the culture medium was 6.0 (Fig. 1A). Optimal activity of cellobiase from Ganoderma australe was obtained after 9 days of solid-state fermentation with an initial pH of 5.5 (Chuwech and Rakariyatham 2014). Maximal activity of β-glucosidase from Aspergillus terreus was also obtained at pH 5.5 (Shahriarinour et al. 2011). A. niger cellulase production reached a maximum at pH 5.0 (Sridevi et al. 2015). The optimal initial pH of fermentation for production of most fungal β-glucosidases ranges from pH 5.0 to pH 6.0, which is approximately consistent with the optimum growth pH of the fungus.
β-glucosidase production during fungal cell growth is usually highly dependent on culture temperature. Therefore, the temperature of M. ardhlaengiktus RSC1 fermentation must be optimized for enzyme production. The MaBgl activity of the liquid medium was assayed after RSC1 was fermented at temperatures ranging from 26 ℃ to 34 ℃, with optimal activity resulting from a fermentation temperature of 30 ℃ (Fig. 1B). Most of the fungal strains producing β-glucosidase have an optimal growth rate at a temperature range of 25 ℃ to 30 ℃ (Kumar et al. 2008). Optimal cellobiose activity from white-rot fungus, Ganoderma austral, was obtained after 9 days of culture at 25 ℃ (Chuwech and Rakariyatham 2014). Maximal production of cellulase (including β-glucosidase) by A. niger MS82 occurred at 35 ℃ under submerged conditions (Sohail et al. 2009).
In general, β-glucosidase production in fungi occurs during a specific growth phase. Maintenance of culture conditions for periods of time longer than necessary would result in avoidable financial costs for industrial scale enzyme production. The MaBgl activity in the liquid medium was assayed at 24-h intervals during 7 days of M. ardhlaengiktus RSC1 fermentation. At each time point, MaBgl activity was assayed at pH 4.8. The MaBgl activity reached a maximum on day 6 (Fig. 1C). Thus, incubation time should be strictly controlled in order to achieve efficient MaBgl production during fermentation of M. ardhlaengiktus RSC1. Kocher et al. (2007) has reported that the activity of β-glucosidase from Trichoderma harzianum was highest after 8 days of incubation.
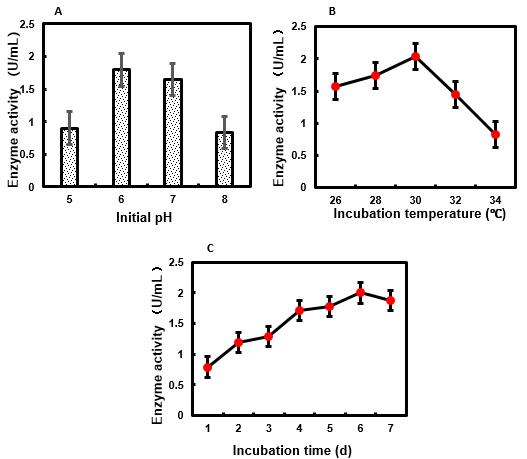
Fig. 1. (A) Effect of initial pH of M. ardhlaengiktus on culture medium on MaBgl production; (B) effect of incubation temperature of M. ardhlaengiktus on MaBgl activity; (C) effect of incubation time of M. ardhlaengiktus on MaBgl activity
Purification and Kinetic Parameters of MaBgl
A two-step protocol was used to purify MaBgl to homogeneity. The crude MaBgl enzyme preparation was precipitated using a range of ammonium sulfate saturation levels from 20% to 80%. Maximal MaBgl activity was achieved with an ammonium sulfate saturation of 20% (Table 1).
Partially purified enzyme from this treatment was used in the further purification of MaBgl. In the purification process of MaBgl, higher enzyme activity can be obtained when ammonia sulfate saturation is 20%. Ammonia sulfate with higher saturation was used to precipitate β-glucosidase in the most of reported fungi (Roy et al. 1991; Zanoelo et al. 2004; Chandra et al.2013; Oh et al. 2018). Oh et al. (2018) reported that the protein precipitates obtained by using 40% or 40 to 60% ammonium sulfate contained 10% and 90% of total activity, respectively. When El-Naggar et al. (2015a) separated β-glucosidase from Aspergillus terreus, β-glucosidase could be obtained by precipitating crude culture filtrate with 20, 40, 60, 80, and 90% ammonia sulfate, but 80% ammonia sulfate precipitation was the best.
Table 1. Effect of Ammonium Sulfate Fractional Precipitation on MaBgl Enzyme Activity
Partially purified MaBgl was concentrated and applied to a DEAE-Sepharose FF anion exchange column. Purified MaBgl was eluted and collected from a single peak with gradient NaCl (Fig. 2A). SDS-PAGE analyses of the purified MaBgl revealed single protein bands corresponding to molecular weight of about 72 kDa for MaBgl (Fig. 2B), which indicated homogeneity of the preparation.
The molecular weights of β-glucosidases produced by different fungi vary. A β-glucosidase with molecular weight of 340 kDa from Aureobasidium pullulans comprised a glycoprotein of two subunits, each with a molecular weight of about 165 kDa (Saha et al. 1994). A. niger β-glucosidase has a molecular weight of approximately 160 kDa (Dan et al. 2000), while the enzyme from P. decumbens presented as a single band of 120 kDa on SDS-PAGE (Chen et al. 2010).
The purification process of MaBgl was started with 100 mL culture filtrate. After precipitation with 20% ammonium sulfate, the recovery of MaBgl enzyme activity was 34.7%, which represents purification by a factor of 3.04 (Table 2). After separation and purification with DEAE-Sepharose FF anion exchange column, the recovery of MaBgl enzyme activity was 5.2%, and the specific activity was increased from 3.15 U/mg to 18.7 U/mg, with a purification of 5.92 times (Table 2). In addition, MaBgl precipitated by 40% ammonium sulfate was not recovered, which will affect the total activity of the enzyme and the recovery rate of the enzyme activity.
Fig. 2. Characteristics of purified MaBgl from M. ardhlaengiktus RSC1: (A) chromatographic profile of MaBgl purified in a DEAE-Sepharose FF column; (B) SDS-PAGE analysis of MaBgl
Table 2. Parameters in MaBgl Purification
Lineweaver-Burk plotting of MaBgl kinetic data yielded the equation y = 2.743x + 35.064 (R2 = 0.989) in Excel 2016 (Fig. 3). The kinetic parameters for the hydrolysis of salicin by the purified enzyme were determined from the equation in Excel 2016. MaBgl exhibited a Michaelis constant (Km) value of 78.2 μmol/L and a maximum velocity (Vmax) of 28.5 μmol/(L·min). The kinetics of many fungal β-glucosidases have been studied (Saha et al. 1994; Chauve et al. 2010; Jeya et al. 2010; Ramani et al. 2012; Kaur et al. 2014), but due to differences in the fungal sources and β-glucosidase substrates used, the kinetic parameters reported in these studies are not directly comparable.
Reaction pH greatly affects enzyme activity, and thus pH should be optimized in order to accurately evaluate enzyme activity. The activity of the crude MaBgl enzyme preparation obtainedfrom M. ardhlaengiktus RSC1 was measured from pH 4.49 to pH 9.18. The MaBgl had maximum activity at pH 4.80 (Fig. 4A). Most fungal β-glucosidases have pH optima in the acidic range (at pH 4.5 to pH 6.0) (Camassola et al. 2004; Chen et al. 2010; Ramani et al. 2012; Santos et al. 2016). The MaBgl activity was also measured at a range of temperatures (30 ℃ to 80 ℃), and its optimum temperature was 50 ℃ (Fig. 4B). At 70 °C, the activity of MaBgl was over 80% of its maximum value, which indicated that MaBgl from M. ardhlaengiktus had considerable thermotolerance. Native β-glucosidases generally have an optimal temperature between 50 ℃ and 65 ℃ (Chen et al. 1992; Kumar et al. 2008; Gusakov 2011), while the optimal temperature of β-glucosidase from Penecillium decumbens was reported to be 65 ℃ to 70 ℃ (Chen et al. 2010).
Fig. 3. Lineweaver-Burk curve of MaBgl kinetic data
Characterization of Enzyme Activity of MaBg1
Fig. 4. (A) Effects of pH on the activity of MaBgl; (B) Effects of temperature on the activity of MaBgl; (C) Thermal stability of MaBgl; (D) pH stability of MaBgl
The residual enzyme activity was determined by holding the enzyme at different temperatures for 0.5 h and 1 h, respectively. The results showed that MaBgl exhibited good thermal stability at 20 and 30 ℃, and retained 93.5% of the enzyme activity at 30 ℃ for 1 h (Fig. 4C). With the increase of temperature and the prolongation of holding time, the residual enzyme activity decreased gradually. When the temperature was higher than 60 ℃, the residual enzyme activity decreased rapidly with the increase of temperature (Fig. 4C). The residual enzyme activity was only 45.2% and 17.3% of the initial enzyme activity after holding for 0.5 h and 1 h at 70 ℃ (Fig. 4C). At 90 ℃, the residual enzyme activity almost disappeared (Fig. 4C), and most proteases had been denatured and inactivated.
The residual enzyme activity was determined by placing the enzyme at 25 ℃ for 1 h and 5 h at different pH. MaBgl was stable between pH 4.6 and 5.0, and the enzyme activity remained basically unchanged (Fig. 4D). When the pH value was 2.2, the residual enzyme activity was only 7.9% and 5.5% when MaBgl were preserved for 1 h and 5 h at 25℃, respectively (Fig. 4D). Later, with the increase of pH, the stability of MaBgl to pH increased. When the pH value is higher than 5.6, the stability of MaBgl to the pH value decreased with the increase of the pH value (Fig. 4D). The residual enzyme activity was only 26.5% and 11.7% when MaBgl were stored at pH8.0 for 1 h and 5 h, respectively (Fig. 4D).
CONCLUSIONS
- This study describes a novel β-glucosidase, MaBgl, produced by the fungal strain M. ardhlaengiktus RSC1 from field rice straw.
- Time-course studies of MaBgl production by the RSC1 strain under optimized conditions revealed that maximum production of β-glucosidase occurred after 6 days of culture. This result indicated the potential usefulness of the enzyme in agricultural and industrial processes, as maintenance of cultures for longer periods of time poses economic costs.
- The optimal pH for MaBgl enzyme activity was 4.80, in the acidic range. The optimal initial pH for the culture medium used for MaBgl production was 6.0, which might be related to the optimal pH for growth of M. ardhlaengiktus RSC1.
- MaBgl was purified to homogeneity by 20% ammonium sulfate saturation and DEAE-Sepharose FF anion exchange column. The molecular weight of native MaBgl was 72 kDa by SDS-PAGE. This molecular weight was similar to that of several other fungal β-glucosidases. The Km and Vmax values of purified MaBgl were 78.2 μmol/L and 28.5 μmol/(L·min), respectively. The study of MaBgl enzymatic properties provides a basis for further development and utilization.
ACKNOWLEDGMENTS
The authors are grateful for the support of the National Natural Science Foundation of China (Grant No. 31760601 and 31360460) and the Key Research and Development Projects in Jiangxi Province (Grant No. 20171BBF60007).
REFERENCES CITED
Ang, S. K., Shaza, E. M., Adibah, Y., Suraini, A. A., and Madihah, M. S. (2013). “Production of cellulases and xylanase by Aspergillus fumigatus SK1 using untreated oil palm trunk through solid state fermentation,” Process Biochem. 48(9), 1293-1302. DOI: 10.1016/j.procbio.2013.06.019
Baldrian, P., and Valášková, V. (2008). “Degradation of cellulose by basidiomycetous fungi,” FEMS Microbiol. Rev. 32(3), 501-521. DOI: 10.1111/j.1574-6976.2008.00106.x
Berghem, L. E. R., Pettersson, L. G., and Fredriksson, U. B. A. (1975). “The mechanism of enzymatic cellulose degradation. Characterization and enzymatic properties of a β-1,4-glucan cellobiohydrolase from Trichoderma viride,” Eur. J. Biochem. 53(1), 55-62. DOI: 10.1111/j.1432-1033.1975.tb04041.x
Bhat, M. K., and Bhat, S. (1997). “Cellulose degrading enzymes and their potential industrial applications,” Biotechnol. Adv. 15(3-4), 583-620. DOI: 10.1016/S0734-9750(97)00006-2
Borgia, P., and Sypherd, P. S. (1977). “Control of beta-glucosidase synthesis in Mucor racemosus,” J. Bacteriol. 130(2), 812-817.
Borgia, P. I., and Mehnert, D. W. (1982). “Purification of a soluble and wall-bound form of beta-glucosidase from Mucor racemosus,” J. Bacteriol. 149(2), 515-522.
Bradford, M. M. (1976). “A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding,” Anal. Biochem. 72(1-2), 248-254. DOI: 10.1016/0003-2697(76)90527-3
Camassola, M., De Bittencourt, L. R., Shenem, N. T., Andreaus, J., and Dillon, A. (2004). “Characterization of the cellulase complex of Penicillium echinulatum,” Biocatal. Biotransform. 22(5-6), 391-396. DOI: 10.1080/10242420400024532
Carlile, M. J., and Watkinson, S. C. (1997). The Fungi, Academic Press, New York, NY, pp. 269-275.
Cavka, A., Alriksson, B., Rose, S. H., van Zyl, W. H., and Jönsson, L. J. (2014). “Production of cellulosic ethanol and enzyme from waste fiber sludge using SSF, recycling of hydrolytic enzymes and yeast, and recombinant cellulase-producing Aspergillus niger,” J. Ind. Microbiol. Biotechnol. 41(8), 1191-1200. DOI: 10.1007/s10295-014-1457-9
Chandra, M., Kalra, A., Sangwan, N. S., and Sangwan, R. S. (2013). “Biochemical and proteomic characterization of a novel extracellular β-glucosidase from Trichoderma citrinoviride,” Mol. Biotechnol. 53(3), 289-299. DOI: 10.1007/s12033-012-9526-7
Chauve, M., Mathis, H., Huc, D., Casanave, D., Monot, F., and Ferreira, N. L. (2010). “Comparative kinetic analysis of two fungal β-glucosidases,” Biotechnol. Biofuels 3(1), 3. DOI: 10.1186/1754-6834-3-3
Chen, H., Hayn, M., and Esterbauer, H. (1992). “Purification and characterization of two extracellular β-glucosidases from Trichoderma reesei,” Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1121(1-2), 54-60. DOI:10.1016/0167-4838(92)90336-C
Chen, M., Qin, Y., Liu, Z., Liu, K., Wang, F., and Qu, Y. (2010). “Isolation and characterization of a β-glucosidase from Penicillium decumbens and improving hydrolysis of corncob residue by using it as cellulase supplementation,” Enzyme Microb. Technol. 46(6), 444-449. DOI: 10.1016/j.enzmictec.2010.01.008
Chuwech, M., and Rakariyatham, N. (2014). “Potential of peanut hulls as substrates for fungal cellulase bioproduction through solid state fermentation,” KKU Res. J. 19, 235-243.
Dan, S., Marton, I., Dekel, M., Bravdo, B.-A., He, S., Withers, S. G., and Shoseyov, O. (2000). “Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger -glucosidase,” J. Biol. Chem. 275(7), 4973-4980. DOI: 10.1074/jbc.275.7.4973
El-Naggar, N. E., Haroun, S. A., ElRazak, A. A., Owis E. A., and Sherie, A. A. (2015a). “Purification and characterization of β-glucosidase produced by Aspergillus terreus under solid state fermentation,” Current Biotechnology 4(3), 380-386. DOI: 10.2174/2211550104999151105095025
El-Naggar, N. E., Haroun, S. A., Oweis, E. A., and Sherief, A. A. (2015b). “Identification of newly isolated Talaromyces pinophilus and statistical optimization of β-glucosidase production under solid-state fermentation,” Prep. Biochem. Biotechnol. 45(7), 712-729. DOI: 10.1080/10826068.2014.943375
Gaur, R., and Tiwari, S. (2015). “Isolation, production, purification and characterization of an organic-solvent-thermostable alkalophilic cellulase from Bacillus vallismortis RG-07,” BMC Biotechnol. 15(19), 1-12. DOI: 10.1186/s12896-015-0129-9
Ghose, T. K. (1987). “Measurement of cellulase activities,” Pure Appl. Chem. 59(2), 257-268. DOI: 10.1351/pac198759020257
Gong, C., Maun, C. M., and Tsao, G. T. (1981). “Direct fermentation of cellulose to ethanol by a cellulolytic filamentous fungus, Monilia sp.,” Biotechnol. Lett. 3(2), 77-82. DOI: 10.1007/BF00145114
Gusakov, A. V. (2011). “Alternatives to Trichoderma reesei in biofuel production,” Trends Biotechnol. 29(9), 419-425. DOI: 10.1016/j.tibtech.2011.04.004
Hernández-Guzmán, A., Flores-Martínez, A., Ponce-Noyola, P., and Villagómez-Castro, J. C. (2016). “Purification and characterization of an extracellular β-glucosidase from Sporothrix schenckii,” FEBS Open Bio 6(11), 1067-1077. DOI: 10.1002/2211-5463.12108
Hmad, I. B., Abdeljalil, S., Saibi, W., Amouri, B., and Gargouri, A. (2014). “Medium initial pH and carbon source stimulate differential alkaline cellulase time course production in Stachybotrys microspora,” Appl. Biochem. Biotechnol. 172(5), 2640-2649. DOI: 10.1007/s12010-013-0705-1
Jeya, M., Joo, A.-R., Lee, K.-M., Tiwari, M. K., Lee, K.-M., Kim, S.-H., and Lee, J.-K. (2010). “Characterization of β-glucosidase from a strain of Penicillium purpurogenum KJS506,” Appl. Microbiol. Biotechnol. 86(5), 1473-1484. DOI: 10.1007/s00253-009-2395-8
Juhász, T., Szengyel, Z., Szijártó, N., and Réczey, K. (2004). “Effect of pH on cellulase production of Trichoderma reesei RUT C30,” Appl. Biochem. Biotechnol. 113(1-3), 201-211. DOI: 10.1385/ABAB:113:1-3:201
Kaur, B., Oberoi, H. S., and Chadha, B. S. (2014). “Enhanced cellulase producing mutants developed from heterokaryotic Aspergillus strain,” Bioresource Technol. 156, 100-107. DOI: 10.1016/j.biortech.2014.01.016
Kiran, E. U., Trzcinski, A. P., Ng, W. J., and Liu, Y. (2014). “Enzyme production from food wastes using a biorefinery concept,” Waste Biomass Valori. 5(6), 903-917. DOI: 10.1007/s12649-014-9311-x
Kocher, G., Kalra, K., and Banta, G. (2007). “Optimization of cellulase production by submerged fermentation of rice straw by Trichoderma harzianum Rut-C 8230,” Internet J. Microbiol. 5(2), 1-5.
Kumar, R., Singh, S., and Singh, O. V. (2008). “Bioconversion of lignocellulosic biomass: Biochemical and molecular perspectives,” J. Ind. Microbiol. Biotechnol. 35(5), 377-391. DOI: 10.1007/s10295-008-0327-8
Lineweaver, H., and Burk, D. (1934). “The determination of enzyme dissociation constants,” J. Am. Chem. Soc. 56(3), 658-666. DOI: 10.1021/ja01318a036
Lynd, L. R., Weimer, P. J., van Zyl, W. H., and Pretorius, I. S. (2002). “Microbial cellulose utilization: Fundamentals and biotechnology,” Microbiol. Mol. Biol. Rev. 66(3), 506-577. DOI: 10.1128/MMBR.66.3.506-577.2002
Nanda, S., Mohammad, J., Reddy, S. N., Kozinski. J. A., and Dalai, A. K. (2014). “Pathways of lignocellulosic biomass conversion to renewable fuels,” Biomass Convers. Biorefin. 4(2), 157-191. DOI: 10.1007/s13399-013-0097-z
Oh, J. M., Lee, J. P., Baek, S. C., Kim, S. G., Jo, Y. D., Kim, J., and Kim, H. (2018). “Characterization of two extracellular β-glucosidases produced from the cellulolytic fungus Aspergillus, sp. ydj216 and their potential applications for the hydrolysis of flavone glycosides,” Int. J. Biol. Macromol. 111, 595-603. Doi:10.1016/j.ijbiomac.2018.01.020
Ortiz, G. E., Guitart, M. E., Cavalitto, S. F., Albertó, E. O., Fernández-Lahore, M., and Blasco, M. (2015). “Characterization, optimization, and scale-up of cellulases production by Trichoderma reesei cbs 836.91 in solid-state fermentation using agro-industrial products,” Bioproc. Biosyst. Eng. 38(11), 2117-2128. DOI: 10.1007/s00449-015-1451-2
Ramani, G., Meera, B., Vanitha, C., Rao, M., and Gunasekaran, P. (2012). “Production, purification, and characterization of a β-glucosidase of Penicillium funiculosum NCL1,” Appl. Biochem. Biotechnol. 167(5), 959-972. DOI: 10.1007/s12010-012-9645-4
Roy, S. K., Raha, S. K., Sadhukhan, R. K., and Chakrabarty, S. L. (1991). “Purification and characterization of extracellular beta-glucosidase from Myceliophthora thermophile,” World J. Microbiol. Biotechnol. 7(6), 613-618. DOI:10.1007/BF00452843
Saha, B. C., Freer, S. N., and Bothast, R. J. (1994). “Production, purification, and properties of a thermostable β-glucosidase from a color variant strain of Aureobasidium pullulans,” Appl. Environ. Microbiol. 60(10), 3774-3780.
Santos, F. R. da S., Garcia, N. F. L., da Paz, M. F., Fonseca, G. G., and Leite, R. S. R. (2016). “Production and characterization of β-glucosidase from Gongronella butleri by solid-state fermentation,” Afr. J. Biotechnol. 15(16), 633-641. DOI: 10.5897/AJB2015.15025
Shahriarinour, M., Wahab, M. N. A., Mohamad, R., Mustafa, S., and Ariff, A. B. (2011). “Effect of medium composition and cultural condition on cellulase production by Aspergillus terreus,” Afr. J. Biotechnol. 10(38), 7459-7467. DOI: 10.5897/AJB11.199
Sherief, A. A., El-Naggar, E. A., and Hamza, S. S. (2010). “Bioprocessing of lignocellulosic biomass for production of bioethanol using thermotolerant aspergillus fumigatus under solid state fermentation conditions,” Biotechnology (Faisalabad), 9(4), 513-522. DOI: 10.3923/biotech.2010.513.522
Sohail, M., Siddiqi, R., Ahmad, A., and Khan, S. A, (2009). “Cellulase production from Aspergillus niger MS82: Effect of temperature and pH,” New Biotechnol. 25(6), 437-441. DOI: 10.1016/j.nbt.2009.02.002
Somkuti, G. A. (1974). “Synthesis of cellulase by Mucor pusillus and Mucor miehei,” J. Gen. Microbiol. 81(1), 1-6. DOI: 10.1099/00221287-81-1-1
Somkuti, G. A., Babel, F. J., and Somkuti, A. C. (1969). “Cellulolysis by Mucor pusillus,” Appl. Microbiol. 17(6), 888-892.
Sridevi, A., Golla, N., Sandhya, A., and Devi, P. S. (2015). “Enhanced production of A. niger cellulase with culture based strategies,” Res. J. Pharm., Biol. Chem. Sci. 6(4), 1042-1049.
Teeri, T. T. (1997). “Crystalline cellulose degradation: New insight into the function of cellobiohydrolases,” Trends Biotechnol. 15(5), 160-167. DOI: 10.1016/S0167-7799(97)01032-9
Tomme, P., Warren, R. A. J., and Gilkes, N. R. (1995). “Cellulose hydrolysis by bacteria and fungi,” Adv. Microb. Physiol. 37, 1-81. DOI: 10.1016/S0065-2911(08)60143-5
Wei, H., Wang, W., Yarbrough, J. M., Baker, J. O., Laurens, L., Van Wychen, S., Chen, X., Taylor, L. E., Xu, Q., Himmel, M. E., and Zhang, M. (2013). “Genomic, proteomic, and biochemical analyses of oleaginous Mucor circinelloides: Evaluating its capability in utilizing cellulolytic substrates for lipid production,” PLoS One 8(9), e71068. DOI: 10.1371/journal.pone.0071068
Wilson, D. B., and Irwin, D. C. (1999). “Genetics and properties of cellulases,” Adv. Biochem. Eng. Biotechnol. 65, 1-21. DOI: 10.1007/3-540-49194-5_1
Wood, T. M., and McCrae, S. I. (1972). “The purification and properties of the C1 component of Trichoderma koningii cellulase,” Biochem. J. 128(5), 1183-1192. DOI: 10.1042/bj1281183
Wood, T. M., and McCrae, S. I. (1977). “Cellulase from Fusarium solani: Purification and properties of the C1 component,” Carbohydr. Res. 57, 117-133. DOI: 10.1016/S0008-6215(00)81925-4
Wood, T. M., and McCrae, S. I. (1986). “The cellulase of Penicillium pinophilum:Synergism between enzyme components in solubilizing cellulose with special reference to the involvement of two immunologically distinct cellobiohydrolases,” Biochem. J. 234(1), 93-99. DOI: 10.1042/bj2340093
Zanoelo, F. F., Maria de Lourdes Teixeira de Moraes Polizeli, Terenzi, H. F., and Jorge, J. A. (2004). “β-glucosidase activity from the thermophilic fungus Scytalidium thermophilum is stimulated by glucose and xylose,” FEMS Microbiol. Lett. 240(2), 137-143. DOI:10.1016/j.femsle.2004.09.021
Zhang, J., Pakarinen, A., and Viikari, L. (2013). “Synergy between cellulases and pectinases in the hydrolysis of hemp,” Bioresource Technol. 129, 302-307. DOI: 10.1016/j.biortech.2012.11.064
Article submitted: October 23, 2018; Peer review completed: December 15, 2018; Revised version received and accepted: January 5, 2019; Published: January 14, 2019.
DOI: 10.15376/biores.14.1.1626-1638
