Abstract
Bacillus amyloliquefaciens xylanase A (baxA), an endoxylanase (EC. 3.2.1.8) gene, was cloned through PCR using the genome of B. amyloliquefaciens as a template. The open reading frame of baxA was 642 bp, and the gene encoded a 213 amino acid protein with a predicted molecular mass of 23.3 kDa. reBaxA1 produced in Escherichia coli with the pET30a(+) vector (T7 lac promoter) formed inclusion body and did not show any xylanase activity. reBaxA2 produced in E. coli with the pCold TF vector (cspA lac promoter) showed high xylanase activity; this enzyme was secreted into the culture medium and remained in the cell. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis analysis showed that the molecular weights of reBaxA1 and reBaxA2 were approximately 28.5 and 77.2 kDa, respectively. The optimal activity of reBaxA2 occurred at 55 °C and pH 6.0. Moreover, the Michaelis–Menten constant (Km) and maximal activity (Vmax) of reBaxA2 were 4.98 mg•ml-1 and 12.79 μmol•min–1•mL–1, respectively. High–performance liquid chromatography analysis showed that reBaxA2 released xylooligosaccharides from birchwood, beechwood, and oat spelt xylans, with xylopentaose, xylotriose, and xylotetraose as major products, respectively.
Download PDF
Full Article
Expression of Recombinant Bacillus amyloliquefaciens Xylanase A in Escherichia coli and Potential Application in Xylan Hydrolysis
Xin Xu,a Mingqi Liu,a,b,* and Xianjun Dai a,b
Bacillus amyloliquefaciens xylanase A (baxA), an endoxylanase (EC. 3.2.1.8) gene, was cloned through PCR using the genome of B. amyloliquefaciens as a template. The open reading frame of baxAwas 642 bp, and the gene encoded a 213 amino acid protein with a predicted molecular mass of 23.3 kDa. reBaxA1 produced in Escherichia coli with the pET30a(+) vector (T7 lac promoter) formed inclusion body and did not show any xylanase activity. reBaxA2 produced in E. coli with the pCold TF vector (cspA lac promoter) showed high xylanase activity; this enzyme was secreted into the culture medium and remained in the cell. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis analysis showed that the molecular weights of reBaxA1 and reBaxA2 were approximately 28.5 and 77.2 kDa, respectively. The optimal activity of reBaxA2 occurred at 55 °C and pH 6.0. Moreover, the Michaelis–Menten constant (Km) and maximal activity (Vmax) of reBaxA2 were 4.98 mg•ml-1 and 12.79 μmol•min–1•mL–1, respectively. High–performance liquid chromatography analysis showed that reBaxA2 released xylooligosaccharides from birchwood, beechwood, and oat spelt xylans, with xylopentaose, xylotriose, and xylotetraose as major products, respectively.
Keywords: Xylanase; Bacillus amyloliquefaciens; Expression; Enzyme properties; Xylooligosaccharides
Contact information: a: Key Laboratory of Marine Food Quality and Hazard Controlling Technology of Zhejiang Province, China JiLiang University, Hangzhou 310018, China; b: National and Local United Engineering Lab of Quality Controlling Technology and Instrumentation for Marine Food, China JiLiang University, Hangzhou 310018, China; *Corresponding author: mqliu524@163.com
INTRODUCTION
Xylan, a poly-pentose polysaccharide, is the major component of hemicelluloses and accounts for one-third of the total plant carbohydrates. Xylan is the second most abundant renewable biological resource in nature, after cellulose (Prade 1995). Xylanases are glycosidases (o-glycoside hydrolases, EC 3.2.1.x) that catalyse the endohydrolysis of 1,4-β-D-xylosidic linkages in xylan (Collins et al. 2005). Xylan can be depolymerised into xyloligosaccharides, xyloses, and oligomers by a complex enzymatic system involving xylanases from all families (Margeot et al. 2009). As xylan contains various side chains, its complete hydrolysis requires the synergistic action of various hydrolytic enzymes, particularly endoxylanases and β-xylosidases (Juturu and Wu 2012). Xylanases exhibit broad application prospects in chemical production, feed and food industries, pulping and laundry industries, and other industries. In the last decade, xylanases have been used to produce tailored xylooligosaccharides (XOS) with prebiotic action for applications in food and feed industries (Pollet et al. 2010). Arabinoxylooligosaccharides or XOS with different substitution and polymerization degrees are obtained after xylanase action on a native biomass substrate (Imaizumi et al. 1991; Swennen et al.2006).
Many bacteria and fungi secrete extracellular xylanases (Kulkarni et al. 1999), but these natural xylanases present several limitations and cannot meet the requirements for industrial production. Xylanases from various strains of Bacillus halodurans and Geobacillus thermoleovorans exhibit activities under alkaline conditions (Mamo et al. 2006; Sharma et al. 2007); however, enzymatic activities at high temperatures are inadequate for industrial process conditions. Xylanases used in several industries must be alkali-stable and thermostable (Verma and Satyanarayana 2012). With the advent of genetic engineering, hundreds of xylanase genes have been cloned and expressed into homologous and heterologous hosts to obtain enzymes for commercial use (Ahmed et al. 2009). The 10A xylanase gene of Saccharomonospora viridis Svixyn was expressed in Escherichia coli, and the recombinant xylanase could not only decrease the viscosity loss of pulp but also improve brightness stability (Wang et al. 2015).
The E. coli expression system is one of the most ideal heterologous protein expression systems. E. colihas been widely used to clone genes and produce recombinant proteins since the 1980s. Several xylanase genes from different sources have been expressed in E. coli (Sriyapai et al. 2011; Elgharbi et al. 2015). The success of this host is attributed to the simplicity of DNA cloning, selectivity of clone vectors, overexpression of recombinant proteins, and easy separation and purification methods (Juturu and Wu 2012). Despite these advantages, active xylanases cannot be usually produced in E. colibecause of the repetitive appearance of rare codons and the requirements for specific post-translational modifications, such as sulphate bond formation (Stewart et al. 1998) and N-glycosylation (Messner 2009). As such, proteins are often synthesized as inclusion bodies, which are unfavourable for separation and industrial applications because of the lack of post-translational modification (Georgiou and Valax 1996). The co-expression of molecular chaperones, which is known to function in the protein-folding pathway in vivo, can increase the solubility of client proteins (Sun et al. 2011; Jhamband Sahoo 2012).
In this study, the gene encoding Bacillus amyloliquefaciens xylanase A (baxA) was cloned and ligated to the pET30a(+) and pCold TF vectors. The baxA was successfully expressed in E. coli BL21 (DE3) under the control of different promoters. The recombinant xylanase (reBaxA2) was purified and characterized, and XOS released from xylans by reBaxA2 was determined and quantified through high-performance liquid chromatography (HPLC).
EXPERIMENTAL
Materials
The B. amyloliquefaciens strain producing xylanase was obtained from the Central Laboratory of Food Science Department, China JiLiang University. The pCold TF DNA cold-shock expression vector and restriction endonucleases were purchased from Takara Biotechnology Co., Ltd. (Dalian). E. coli BL21 (DE3) was used to express baxA. Oat spelt, birchwood, and beechwood xylans were purchased from Sigma. Primers were synthesised at Shanghai Sunny Biotechnology Co., Ltd. Medium components were obtained from Oxoid, whereas xylose (X) from Merck. Standard XOS (xylobiose, X2; xylotriose, X3; xylotetraose, X4; xylopentaose, X5; and xylohexaose, X6) were acquired from Megazyme. All other chemicals were of analytical grade.
Methods
Cloning of baxA and construction of expression plasmid
The B. amyloliquefaciens genome was extracted according to the directions in the general AllGen kit (Beijing Cowin Biotech Co., Ltd). The full-length of baxA was amplified from B. amyloliquefaciens by using the Expand High Fidelity PCR system with a pair of gene specific primers: P1: 5′-GGGTACCATGTTTAAGTTTAAAAAG-3′ and P2: 5′-AAGCTTCCACACTGTTACGTTTGAAC-3′ containing KpnI and HindIII restriction sites (underlined), respectively. The reaction was conducted using ‘touchdown’ PCR under the following conditions: initial denaturation at 95 °C for 5 min; followed by 32 consecutive cycles of denaturation at 94 °C for 30 s, annealing for 50 s at 52 °C with a 0.3 °C decline per cycle and extension at 72 °C for 1 min; and final extension at 72 °C for 10 min. PCR product was purified using a gel extraction kit (Omega Biotechnology Co., USA). After digestion with KpnI and HindIII, the purified product was inserted into the pET30a(+) and pCold TF vectors, respectively. The sequences of the recombinant expression plasmids pCold-baxA and pET-baxA were confirmed through sequencing.
Expression of baxA in E. coli BL21 (DE3)
The pCold-baxA and pET-baxA plasmids were each transformed into E. coli BL21 (DE3) (Sambrookand Russel 2001). The positive clones were identified on Luria–Bertani (LB) agar plates containing appropriate antibiotics and then confirmed through PCR. Positive transformants with relatively high xylanase activity in small-culture analysis were selected for scale-up expression.
No colony showed any xylanase activity in the fermentation and ultrasonicated supernatants among 210 transformants harboring the pET-baxA plasmid. pEbaxA7 was used for sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blot analysis; xylanase produced by this transformant was defined as reBaxA1. The transformant, pCbaxA15, containing the pCold-baxA plasmid showed the highest xylanase activity, and the produced xylanase was designated as reBaxA2. After extraction and purification, reBaxA2 was used for enzyme activity measurements. The pCbaxA15 was then cultured in 250 mL of LB medium with 100 μg/mL ampicillin in a 2L baffled shake flask at 37 °C rotated at 150 rpm. When the bacteria density reached OD600=0.5, the baffled shake flask was incubated at 15 °C for 30 min. Isopropyl-b-D-thiogalactopyranoside (IPTG) was added to the medium at a final concentration of 1.0 mM to induce reBaxA2 production, and the cells were cultured and shaken for 24 h at 15 °C. Microbial optical density (OD) at 600 nm and xylanase activity were determined every 6 h.
Extraction and purification of reBaxA2
The fermentation supernatant of pCbaxA15 was collected through centrifugation at 10,000×g for 15 min at 4 °C. The cell pellet was resuspended in PBS buffer (pH 7.4) and sonicated twice (on for 3 s, off for 5 s, and working time of 5 min) in an ice bath. After centrifugation at 10,000×g for 15 min at 4 °C, the ultrasonicated supernatant and insoluble fractions were separated.
The ultrasonicated supernatant was mixed with high-affinity Ni2+-charged resin (GenScript Corporation) and incubated for 2 h by gently inverting in an ice bath to bind the protein to the resin. The slurry was transferred to a column, and the column was washed with 8 bed volumes of wash buffer and 10 bed volumes of elution buffer. Eluate was collected step by step for subsequent analysis.
Recombinant xylanase activity assay
Xylanase activity was determined using the 3,5-dinitrosalicylic acid procedure with D-xylose as the standard (Miller 1959) and 0.5% (w/v) beechwood xylan as the substrate. Protein concentration was measured using the dye-binding method of Bradford with bovine serum albumin as the standard (Bradford 1976). One unit of enzyme activity was defined as the amount that released 1 μmol reducing sugar equivalents per minute from the substrate under the assay conditions. For each assay, triplicate measurements were conducted to obtain the mean activity value.
Kinetic parameters, including the maximum reaction rate (Vmax) and Michaelis–Menten constant (Km) of reBaxA2, were determined from initial velocities using beechwood xylan as the substrate (2, 3, 4, 5, 6, 7, 8, 10, and 15 mg/mL). Furthermore, the catalytic activities of purified reBaxA2 were measured using oat spelt, beechwood, and birchwood xylans at 55 °C and pH 6.0.
SDS-PAGE and western blot analysis of recombinant xylanases
All the samples from the two types of transformants were subjected to SDS-PAGE. In the Laemmli system, stacking and separating gels consisted of 5% and 12% polyacrylamide, respectively (Laemmli 1976). Proteins were visualised in the gel through Coomassie blue R-250 staining. For western blot analysis, the proteins were transferred to a polyvinylidene difluoride membrane. The membrane was incubated with anti-His monoclonal antibody. Immunoreactive protein was visualised using horseradish peroxidase-labelled goat anti-mouse IgG as the secondary antibody on a chromogenic substrate.
Optimum temperature and thermostability of reBaxA2
The optimum temperature of reBaxA2 was determined through incubation with beechwood xylan dissolved in deionised water at different temperatures (30 °C to 90 °C). The thermal stability of reBaxA2 was studied by measuring the remaining activity after enzyme incubation at different temperatures (30 °C to 90 °C) for 1 h.
Optimum pH and pH stability of reBaxA2
The effect of pH on xylanase activity was measured over a pH range of 3.0 to 8.0 (Na2HPO4–citric acid buffer) and pH 9.0 (0.2 M glycine and 0.2 M NaOH buffer system) at 55 °C. The pH stability of reBaxA2 was determined after incubation in buffers with pH ranging from 3.0 to 9.0 at 25 °C for 1 h, and residual activity was determined at 55 °C and pH 6.0.
Effect of metal ions on reBaxA2 activity
Solutions of metal ions (Zn2+, Cu2+, Co2+, Mg2+, Mn2+, Ca2+, Al3+, and K+) were added to reBaxA2 to obtain a final concentration of 5 mM, and the mixture was incubated at 25 °C for 1 h. The residual activity of reBaxA2 was detected under the optimal conditions.
Hydrolysis products from xylans by reBaxA2
Briefly, reBaxA2 (0.35 U) was incubated with 0.5 mL of beechwood, birchwood, or oat spelt xylans (1.0%, w/v) at 55 °C for 10 min. The samples were boiled for 10 min to terminate the enzymatic hydrolysis and then immediately cooled on ice. The samples were centrifuged at 13000×g and 4 °C for 10 min. The supernatant was analysed through HPLC with a Sugar-pak™ 1 column (300 mm length and 6.5 mm diametre, Waters) by using pure water as the mobile phase (0.5 mL/min) under injection volumes of 20 μL. The column was maintained at 85 °C. Sugar peaks were screened using a Waters 2410 refractive index detector (Liu et al. 2013). Xylose and XOS (X2 to X6) were used as standards.
Nucleotide
The nucleotide sequence for baxA was deposited in the GenBank database under accession number KM 624029.
RESULTS AND DISCUSSION
Amplification and Expression of baxA in E. coli
The baxA gene amplified from the B. amyloliquefaciens genome was separated and recovered through electrophoresis (Fig. 1). The size of the PCR product was 642 bp as determined through sequencing. The product encoded an open reading frame with 213 amino acid residues, including a 28 amino acid N-terminal signal peptide, 2 catalytic sites, and 6 potential N-glycosylation sites as determined through analysis with the NetNGlyc 1.0 Server. The predicted molecular mass of the deduced protein was 23.3 kDa. The nucleotide sequences of baxA and the deduced amino acid sequences are shown in Fig. 2. BLAST analysis showed that baxA was 99% similar to the endo-1,4-beta-xylanase genes from Paenibacillus macerans, Bacillus subtilis, and B. amyloliquefaciens (Accession numbers: AAZ17386.1, WP003151206.1 and WP012118650.1, respectively).
Fig. 1. PCR amplification of baxA; M: DNA Ladder; and Lane 1: PCR product
Fig. 2. Nucleotide sequences of baxA and the deduced amino acid sequences. Note: The signal peptide is underlined, the two catalytic residues are indicated with boxes, the potential N-glycosylation sites are in grey boxes and the stop codon is indicated with an asterisk.
Production and Purification of Recombinant Xylanases
After adding IPTG, the pEbaxA7 was cultivated at 30 °C for 8 h. The fermentation and ultrasonication supernatants did not show any xylanase activity. The reBaxA1 formed inclusion bodies.
The production of reBaxA2 was induced by the addition of IPTG to the medium and incubation at 15 °C. The pCold TF vector provides cold shock technology for high-yield protein expression combined with trigger factor (chaperone) expression to facilitate correct protein folding, thereby enabling efficient soluble protein production for intractable target proteins. The reBaxA2 was secreted into the extracellular matrix and in the cell and was significantly purified after Ni2+-chelating affinity chromatography. The enzyme activities of purified reBaxA2 were 2.63, 3.68, and 3.16 U•mg−1 for beechwood, birchwood, and oat spelt xylans, respectively. The results showed that reBaxA2 exhibited the highest substrate selectivity for birchwood xylan. According to a recent research, the different molecular structures of the substrates could lead to inconsistencies in the measured enzyme activities; in this study, enzyme activities significantly differed when oat and beechwood xylans were used as substrates (Deng et al. 2006). In comparison, a thermostable xylanase from Thermomyces lanuginosusshowed activities in the extracellular matrix (36±1.3 U•mL−1) and the periplasm (42±4 U•mL−1) (Le et al. 2014). Moreover, Thermotoga thermarum produced xylanase with a specific activity of 148.5 U•mg−1 (Shi et al. 2013). Xylanase from Aspergillus niger showed a specific activity of 808.5 U•mg-1towards 1% (w/v) of birchwood xylan (Do et al. 2013).
The activities of reBaxA2 for various beechwood xylan concentrations were plotted in Lineweaver–Burk graphs. Vmax and Km were then calculated. The Km and Vmax of reBaxA2 were 4.98 mg•mL−1and 12.79 μmol•min−1•mL−1, respectively.
Effect of Different Induction Times on pCbaxA15 growth and reBaxA2 Production
The pCold TF vector used “cold-shock” technology. When the temperature decreased from 37 °C to 15 °C, the production of other cellular proteins was suppressed, thereby temporarily halting the overall cell growth. Figure 3 reveals that the microbial OD of pCbaxA15 reached the maximum 12 h after induction, after which the growth slightly fluctuated until 36 h. Furthermore, pCbaxA15 enhanced reBaxA2 synthesis, reached the maximum activity at 24 h and then slowly decreased. Therefore, the optimal induction time was 24 h for reBaxA2 production.
Fig. 3. Effect of induction time on pCbaxA15 growth and reBaxA2 production
SDS-PAGE and Western Blot Analysis of Recombinant Xylanases
The baxA gene encoded a protein with a molecular mass of 23.3 kDa, and reBaxA1 was produced as inclusion bodies under the control of the T7 lac promoter. SDS-PAGE and western blot analysis showed that the molecular mass of reBaxA1, including the protein encoded by baxA, as well as two His-tags and S-tag of the pET30a(+) vector, was about 28.5 kDa (Fig. 4). The reBaxA2 produced under the control of the cspA lac promoter was combined with the trigger factor (54 kDa). The production of the target protein (77.2 kDa) was verified using the pCold TF vector through SDS-PAGE electrophoresis and western blot analysis; a part of the recombinant protein was insoluble (Fig. 5).
Fig. 4. Analysis of reBaxA1 through SDS-PAGE (a) and western blot analysis (b). Note: Lane M: standard protein marker; Lane 1: induced E. coli BL21 (DE3) harboring the pET30a(+) vector; and Lane 2: pEbaxA7 induced by IPTG
Fig. 5. Analysis of reBaxA2 through SDS-PAGE (a) and western blot analysis (b). Note: Lane M: standard protein marker; Lane 1: purified reBaxA2 by Ni2+-chelating affinity chromatography; Lane 2: ultrasonicated supernatant of pCbaxA15 induced by IPTG; Lane 3: fermentation supernatant of pCbaxA15 induced by IPTG; Lane 4: pCbaxA15 induced by IPTG; and Lane 5: E. coli BL21 harboring the pCold TF vector
Optimum Temperature and Thermostability of reBaxA2
The reBaxA2 activity increased with increasing temperature, reached a maximum at 55 °C, and then decreased with further increase in temperature (Fig. 6). The relative activity was higher than 80% between 40 °C and 55 °C. The reBaxA2 was stable at 50 °C for 1 h, and the residual activity was higher than 60%. As the temperature increased, while holding incubation time constant at 1 h, the residual activity rapidly decreased and reached less than 20% at temperature higher than 55 °C (Fig. 6).
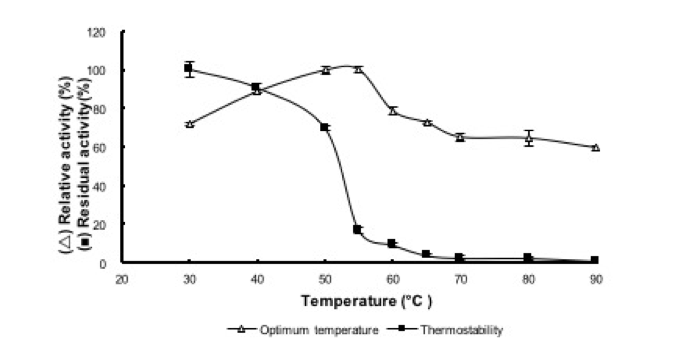
Fig. 6. Effect of temperature on activity and stability
of reBaxA2
Optimum pH and pH Stability of reBaxA2
Enzyme activity was determined using xylan substrates with different pH values. The reBaxA2 showed high activity within a pH range of 4.0 to 7.0, in which relative activity remained higher than 80%. The optimum pH was 6.0 (Fig. 7), and the enzyme was stable from pH 4.0 to 8.0. More than 60% of xylanase activity was retained after incubation under a certain pH range of 4.0 to 8.0 for 1 h at room temperature.
Fig. 7. Effect of pH on activity and stability
of reBaxA2
Effect of Metal Ions on reBaxA2 Activity
The effect of metal ions on reBaxA2 activity was determined by adding Mn2+, Ca2+, Al3+, K+, Zn2+, Cu2+, Co2+, or Mg2+ to the enzyme to a final concentration of 5 mM at 25 °C for 1 h. The reBaxA2 added with pure water was designated as the control. Figure 8 shows that all the eight metal ions could activate recombinant xylanase activity, with Co2+ exhibiting the highest (best) activation performance.
Fig. 8. Effects of metal ions on reBaxA2 stability
Hydrolysis Products from Xylans by reBaxA2
Xylans from oat spelt, beechwood, and birchwood were incubated with reBaxA2 to determine the hydrolysis characteristics of reBaxA2. Hydrolysis products were quantitatively analysed through HPLC (Figs. 9b–d). Xylose and XOS (X2 to X6) were also analysed through HPLC (Fig. 9a).
The hydrolysis products of the three xylans were X2 to X5 (Figs. 9b–d, Table 1). X4 (58.98%), X3 (36.79%), and X5 (38.66%) were the major products for oat spelt, beechwood, and birchwood xylans with concentrations of 1.764, 1.168, and 0.775 mg/mL, respectively. The concentrations of hydrolysis products from oat spelt xylan were lower than those from beechwood xylan, which could be due to the low solubility of oat spelt xylan. Meanwhile, the polymerization degree of main hydrolysis products from oat spelt xylan was higher than that from beechwood xylan, i.e., X4 and X5 compared with X3. These results indicated that beechwood xylan yielded the highest amount of hydrolysis product.
Xylanases with different substrate specificities produce different end products. Some xylanases can degrade xylan to X and X2, whereas others can only produce large oligosaccharides (Reilly 1981). The hydrolysis profiles of beechwood xylan were similar to those of birchwood xylan. Meanwhile, beechwood xylan was more hydrolysed and differed from oat spelt xylan, which may be the consequence of different substrate compositions. Beechwood and birchwood (hardwood) xylans contain o-acetyl-4-o-methylglucuronoxylans and are highly acetylated at C-3 and C-2 positions. The presence of these acetyl groups is responsible for the partial solubility of xylan in water, which could explain the higher solubility of beechwood and birchwood xylans than that of oat spelt xylan. Oat spelt xylan, which fall into the same class as softwood xylans, contain arabino-4-o-methylglucuroxylans. Instead of acetyl groups, oat spelt xylan contains arabinofuranose units (Beg et al. 2001). All these results suggested that reBaxA2 exhibited substrate specificity, and hydrolysis modes varied because of different substrate compositions. In addition, reBaxA2 exhibited a good activity against xylan polymers containing acetyl groups and arabinose substituents of the xylose main chain.
The hydrolysis characteristics of reBaxA2 can be potentially utilised to produce XOS, which is a promising prebiotic compound because of its abundance, inexpensive sources, and various beneficial physiological effects. XOS shows favourable technological features, including stability in acidic media, resistance to heat, and provision to achieve low available energy and significant biological effects at low daily intakes (Moure et al. 2006). Currently, several non-utilised and under-utilised agricultural by-products, including corn cobs (Samanta et al. 2012), wheat straw, and rice husks (Nabarlatz et al. 2007), have been used to obtain XOS through enzymatic hydrolysis. Utilising these wastes can provide a suitable disposal method and thus confer economic and ecological benefits. Imaizum et al. (1991) reported that XOS improved diabetic symptoms, including reduction of liver triglycerides and phosphatidylcholine. XOS also exhibited a probiotic effect by selectively simulating the growth of beneficial intestinal bacteria, such as Bifidobacterium and Lactobacillus, when consumed as a part of diet (Manisseri and Gudipati 2012). A controlled administration of XOS may restrain the growth of pathogenic bacteria, retard disorders caused by imbalanced fermentation in colon, and avoid intestinal disorders, such as constipation, inflammatory bowel disease, diarrhea, and gastritis (Moure et al. 2006). XOS also demonstrated various physiological activities such as promoting immune status, reducing cholesterol, improving the biological availability of calcium, and producing short chain fatty acids (Akpinar et al. 2009). XOS can also effectively ameliorate oxidative stress. Furthermore, reBaxA2 can be applied to convert plant lignocellulose biomass into fermentable sugars; this technology can be used to produce sustainable biofuels in biorefineries, in which agricultural wastes may be utilised for direct bioconversion into chemicals and fuels (Turner et al. 2007).
Fig. 9. HPLC profiles of xylan degradation products. Note: The positions of xylose (1), xylobiose (2), xylotriose (3), xylotetraose (4), xylopentaose (5) and xylohexaose (6) are shown (a). HPLC analysis of hydrolysis products from different xylans by reBaxA2 (b): oat spelt xylan; (c): beechwood xylan; and (d): birchwood xylan
Table 1. Concentration and Percentage Content of Hydrolysis Products from Xylans by reBaxA2
CONCLUSIONS
- The baxA gene was successfully cloned and expressed in E. coli under the control of different promoters (cspA lac and T7 lac).
- The reBaxA1 produced in E. coli under the control of the T7 lac promoter formed inclusion bodies and did not show any activity. The reBaxA2 produced in E. coli under the control of the cspA lac promoter showed high xylanase activity. The cspA lac promoter and co-expression of the trigger factor chaperone effectively increased the amounts of soluble reBaxA2.
- The reBaxA2 generated a series of XOS (X2 to X5) from birchwood, beechwood, and oat spelt xylans and demonstrated a potential for industrial production of XOS.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 31201831) and the Zhejiang Provincial Natural Science Foundation of China (No. LY15C200013). We thank Dr. Shang-Wei Chen for his kind assistance in the HPLC analysis.
REFERENCES CITED
Ahmed, S., Riaz, S., and Jamil, A. (2009). “Molecular cloning of fungal xylanases: An overview,” Appl. Microbiol. Biotechnol. 84, 19-35. DOI: 10.1007/s00253-009-2079-4
Akpinar, O., Erdogan, K., and Bostanci, S. (2009). “Enzymatic production of xylooligosaccharide from selected agricultural wastes,” Food Biopro. Process. 87, 145-151. DOI: 10.1016/j.fbp.2008.09.002
Beg, Q. K., Kapoor, M., Mahajan, L., and Hoondal, G. S. (2001). “Microbial xylanases and their industrial applications: A review,” Appl. Microbiol. Biotechnol. 56, 326-338. DOI: 10.1007/s002530100704
Bradford, M.M. (1976). “A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding,” Anal. Biochem. 72, 248-254. DOI: 10.1016/0003-2697(76)90527-3
Collins, T., Gerday, C., and Feller, G. (2005). “Xylanase, xylanase families and extremophilic xylanases,” FEMS Microbiol. Rev. 29, 3-23. DOI: 10.1016/j.femsre.2004.06.005
Deng, P., Cao, Y. H., Lu, W. Q., and Guo, X. H. (2006). “Expression of xylanase gene xynB fromAspergillus niger in Escherichia coli and characterization of its recombinant xylanase,” Journal of Agricultural Biotechnology. 14(5), 774-778.
Do, T. T., Quyen, D. T., Nguyen, T. N., and Nguyen, V. T. (2013). “Molecular characterization of a glycosyl hydrolase family 10 xylanase from Aspergillus niger,” Protein Expres. Purif. 92, 196-202. DOI: 10.1016/j.pep.2013.09.011
Elgharbi, F., Hlima, H. B., Khemakhem, A. F., Zouari, D. A., Bejar, S., and Sayari, A. H. (2015). “Expression of A. niger US368 xylanase in E. coli: Purification, characterization and copper activation,” Int. J. Biol. Macromol. 74, 263-270. DOI: 10.1016/j.ijbiomac.2014.12.005
Georgiou, G., and Valax, P. (1996). “Expression of correctly folded proteins in Escherichia coli,” Curr. Opin. Biotechnol. 7(2), 190-197. DOI: 10.1016/S0958-1669(96)80012-7
Imaizumi, K., Nakatsu, Y., Sato, M., Sedarnawati, Y., and Sugano, M. (1991). “Effects of xylooligosaccharides on blood glucose, serum and liver lipids and cecum short-chain fatty acids in diabetic rats,” Agric. Biol. Chem. 55, 199-205. DOI: 10.1271/bbb1961.55.199
Juturu, V., and Wu, J. C. (2012). “Microbial xylanases: Engineering, production and industrial applications,” Biotechnol. Adv. 30, 1219-1227. DOI: 10.1016/j.biotechadv.2011.11.006
Jhamb, K., and Sahoo, D. K. (2012). “Production of soluble recombinant proteins in Escherichia coli: Effects of process conditions and chaperone co-expression on cell growth and purification of xylanase,” Bioresour. Technol. 123, 135-143. DOI: 10.1016/j.biortech.2012.07.011
Kulkarni, N., Shendye, A., and Rao, M. (1999). “Molecular and biotechnological aspects of xylanases,” FEMS Microbiol. Rev. 23, 411-456. DOI: 10.1111/j.1574-6976.1999.tb00407.x
Laemmli, U. K. (1976). “Cleavage of structural proteins during the assembly of the head of bacteriophage T4,” Nature 227, 680-685. DOI: 10.1038/227680a0
Le, Y. L., and Wang, H. L. (2014). “High-level soluble expression of a thermostable xylanase from thermophilic fungus Thermomyces lanuginosus in Escherichia coli via fusion with OsmY protein,” Protein Expres. Purif. 99, 1-5. DOI:10.1016/j.pep.2014.03.004
Liu, M. Q., Dai, X. J., Liu, G. F., and Wang, Q. (2013). “Obtaining cellulose binding and hydrolyzing activity of a family 11 hybrid xylanase by fusion with xylan binding domain,” Protein Expres. Purif.88, 85-92. DOI: 10.1016/j.pep.2012.11.014
Mamo, G., Delgado, Martinez, O. A., Mattiasson, B., and Kaul, R. H. (2006). “Cloning, sequencing analysis, and expression of a gene encoding an endoxylanase from Bacillus halodurans S7,” Mol. Biotechnol. 33, 149-159. DOI: 10.1385/MB:33:2:149
Manisseri, C., and Gudipati, M. (2012). “Prebiotic activity of purified xylobiose obtained from ragi (Eleusine coracana, Indaf-15) bran,” Indian J. Microbiol. 52, 251-257. DOI: 10.1007/s12088-011-0176-4
Margeot, A., Hahn-Hagerdal, B., Edlund, M., Slade, R., and Monot, F. (2009). “New improvements for lignocellulosic ethanol,” Curr. Opin. Biotechnol. 20, 372-80. DOI: 10.1016/j.copbio.2009.05.009
Messner, P. (2009). “Prokaryotic protein glycosylation is rapidly expanding from “curiosity” to “ubiquity,” Chem. Bio. Chem. 10, 2151-2154. DOI: 10.1002/cbic.200900388
Miller, G. L. (1959). “Use of dinitrosalicylic acid reagent for determination of reducing sugar,” Anal. Biochem. 31, 426-428. DOI: 10.1021/ac60147a030
Moure, A., GullÓn, P., DomÍnguez, H., and ParajÓ, J. C. (2006). “Advances in the manufacture, purification and applications of xylo-oligosaccharides as food additives and nutraceuticals,” Process Biochem. 41, 1913-1923. DOI: 10.1016/j.procbio.2006.05.011
Nabarlatz, D., Ebringerova, A., and Montane, D. (2007). “Autohydrolysis of agricultural byproducts for the production of xylooligosaccharides,” Carbohydr. Polym. 69, 20-28. DOI: 10.1016/j.carbpol.2006.08.020
Pollet, A., Delcour, J. A., and Courtin, C. M. (2010). “Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families,” Crit. Rev. Biotechnol. 30, 176-91. DOI: 10.3109/07388551003645599
Prade, R. A. (1995). “Xylanase: From biology to biotechnology,” Biotech. Genet. Eng. Rev. 13, 100-131. DOI: 10.1080/02648725.1996.10647925
Reilly, P. J. (1981). “Xylanases: Structure and function,” Trends in the Biology of Fermentation for Fuels and Chemicals, A. Hollaender (ed.), Plenum Press, New York. DOI: 10.1007/978-1-4684-3980-9_8
Samanta, A. K., Senani, S., Kolte, A. P., Sridhar, M., Sampath, K. T., Jayapal, N., and Devi, A. (2012). “Production and in vitro evaluation of xylooligosaccharides generated from corn cobs,” Food Bioprod. Process. 90, 466-474. DOI: 10.1016/j.fbp.2011.11.001
Sambrook, J., and Russel, D. W. (2001). “Plasmids and their usefulness in molecular cloning,” Molecular Cloning: A laboratory manual (Third edition), J. Argentine (ed.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Sharma, A., Adhikari, S., and Satyanarayana, T. (2007). “Alkali-thermostable and cellulose-free xylanase production by an extreme thermophile Geobacillus thermoleovorans,” World J. Microbiol. Biotechnol. 23, 483-490. DOI: 10.1007/s11274-006-9250-1
Shi, H., Zhang, Y., Li, X., Huang, Y. J., Wang, L. L., Wang, Y., and Ding, H. H. (2013). “A novel highly thermostable xylanase stimulated by Ca2+ from Thermotoga thermarum: cloning, expression and characterization,” Biotechnol. Biofuels. 6(26). DOI: 10.1186/1754-6834-6-26
Sun, T., Shen, N., Bai, Y., Li, W. H., and Wei, P. (2011). “Fusion expression of an extreme-thermostable xylanase B64 gene from Thermotoga maritima MSB8 in Escherichia coli,” Microbiology China 38(7), 1090-1097.
Sriyapai, T., Somyoonsap, P., Matsui, K., Kawai, F., and Chansiri, K. (2011). “Cloning of a thermostable xylanase from Actinomadura sp. S14 and its expression in Escherichia coli and Pichia pastoris,” J. Biosci. Bioeng. 111, 528-536. DOI: 10.1016/j.jbiosc.2010.12.024
Stewart, E. J., Åslund, F., and Beckwith, J. (1998). “Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins,” EMBO J. 17, 5543-5550. DOI: 10.1093/emboj/17.19.5543
Swennen, K., Courtin, C. M., and Delcour, J. A. (2006). “Non-digestible oligosaccharides with prebiotic properties,” Crit. Rev. Food Sci. Nutr. 46, 459-71. DOI: 10.1080/10408390500215746
Turner, P., Mamo, G., and Karlsson, E. N. (2007). “Potential and utilization of thermophiles and thermostable enzymes in biorefining,” Microb. Cell Fact. 6, 9-32. DOI: 10.1186/1475-2859-6-9
Verma, D., and Satyanarayana, T. (2012). “Molecular approaches for ameliorating microbial xylanases,” Bioresour. Technol. 117, 360-367. DOI: 10.1016/j.biortech.2012.04.034
Wang, J. F., Wu, Y. Y., Xie, X. M., and Yang, M. (2015). “Effects of xylanase expressed and secreted by green sugar spore bacteria xylanase Svixyn 10A gene in different strains on pre-treating of pulp,” Paper and Paper Making. 34, 37-39.
Article submitted: April 6, 2015; Peer review completed: May 29, 2015; Revisions accepted: June 9, 2015; Published: June 12, 2015.
DOI: 10.15376/biores.10.3.4694-4711
