Abstract
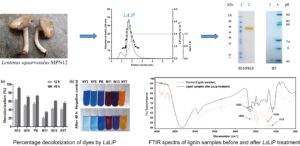
Lignin peroxidase (LiP), which has been studied extensively in white-rot basidiomycetes and their potential to degrade dyes from textile wastewater, plays a role in the biodegradation of lignin from pulp and paper industry wastewater, as well as agricultural waste. Lignin peroxidase (LsLiP) was successfully purified from the newly isolated Lentinus squarrosulus MPN12 with a 47.1-fold purification and a 15.7% yield. After 48 h-incubation, LsLiP was able to decolorize all tested dyes up to 92% for Acid Blue 62 (NY3), followed by Porocion Brilliant blue HGR (PB, 73.5%), Acid Blue 281 (NY5, 70.5%), Acid Blue 113 (IN13, 61%), Acid red 266 (NY7, 56%), and 34.5% for Acid red 299 (NY1), compared to the negative control with the heat-denatured enzyme. The biodegradation potential of LsLiP was further suggested by the change of lignin structure based on Fourier transform infrared (FTIR) analyses. Lignin structure was noticeably changed before and after LsLiP treatment, especially in the fingerprint regions (1600 to 1000 cm-1) and 2930 cm-1 corresponding to the stretching vibrations of various groups in lignin. Thus, LsLiP has potential application in both enzyme-based decolorization of synthetic dyes and lignin biodegradation.
Download PDF
Full Article
Lignin Peroxidase from the White-rot Fungus Lentinus squarrosulus MPN12 and its Application in the Biodegradation of Synthetic Dyes and Lignin
Vu Dinh Giap,a,c Do Huu Nghi,a,b,* Le Huu Cuong,b and Dang Thu Quynh a,b,*
Lignin peroxidase (LiP), which has been studied extensively in white-rot basidiomycetes and their potential to degrade dyes from textile wastewater, plays a role in the biodegradation of lignin from pulp and paper industry wastewater, as well as agricultural waste. Lignin peroxidase (LsLiP) was successfully purified from the newly isolated Lentinus squarrosulus MPN12 with a 47.1-fold purification and a 15.7% yield. After 48 h-incubation, LsLiP was able to decolorize all tested dyes up to 92% for Acid Blue 62 (NY3), followed by Porocion Brilliant blue HGR (PB, 73.5%), Acid Blue 281 (NY5, 70.5%), Acid Blue 113 (IN13, 61%), Acid red 266 (NY7, 56%), and 34.5% for Acid red 299 (NY1), compared to the negative control with the heat-denatured enzyme. The biodegradation potential of LsLiP was further suggested by the change of lignin structure based on Fourier transform infrared (FTIR) analyses. Lignin structure was noticeably changed before and after LsLiP treatment, especially in the fingerprint regions (1600 to 1000 cm-1) and 2930 cm-1 corresponding to the stretching vibrations of various groups in lignin. Thus, LsLiP has potential application in both enzyme-based decolorization of synthetic dyes and lignin biodegradation.
DOI: 10.15376/biores.17.3.4480-4498
Keywords: Lignin peroxidase; Lentinus squarrosulus; Biodegradation; Synthetic dyes; Lignin
Contact information: a: Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, 10000, Hanoi, Vietnam; b: Institute of Natural Products Chemistry, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, 10000, Hanoi, Vietnam; c: HaUI Institute of Technology, Hanoi University of Industry (HaUI), 298 Cau Dien, Bac Tu Liem, 10000, Hanoi, Vietnam; *Corresponding author: quynhdang1994@gmail.com; dohnghi@gmail.com
GRAPHICAL ABSTRACT

INTRODUCTION
Lignin is a natural macromolecule having a cross-linked polyphenol structure (Garguiak and Lebo 2000). The complex polymer structure contains phenolic hydroxyl groups, methoxy groups, and some terminal aldehyde groups in the side chains. The aromatic rings are abundantly present in lignin in the form of the basic phenylpropane units, which are syringyl, guaiacyl, and p-hydroxyphenyl (Zhang and Naebe 2021). They are responsible for maintaining the rigidity of the plant’s cell wall. Thus, lignin is known to provide protection to plants against microbial and enzymatic attacks. With a persistent structure and difficulty in biodegradation, the presence of lignin and its derivatives in industries increases the pollution load on the environment. Indeed, lignin is present in abundance in pulp and paper industry wastewater, and agricultural waste (wheat straw, rice straw, and bagasse), etc. (Gaur et al. 2018; Pandit et al. 2021). Therefore, degrading lignin to reduce its negative impact on the environment is an urgent problem.
Lignin peroxidase (LiP, EC 1.11.1.14) is a ligninolytic enzyme, including manganese peroxidase (MnP, EC 1.11.1.13), lignin peroxidase, versatile peroxidases (VeP, EC 1.11.1.16), and laccase (Lac, EC 1.10.3.2). They are enzymes of biotechnological interest due to their ability to oxidize high redox potential aromatic compounds, including the lignin polymer. Lignin peroxidase was first discovered in Phanerochaete chrysosporium by Tien and Kirk (1983). It catalyzes the oxidative characteristics of a variety of non-phenolic and phenolic lignin aromatic complex compounds in the presence of H2O2 (Tien and Kirk 1984; Hamid and Rehman 2009), resulting in the cleavage of the Cɑ-Cβ bond, aromatic ring opening, and phenolic oxidation (Christian et al. 2005).
Due to its high redox potential and enlarged substrate range, LiP is great for application in various industrial fields. For instance, LiP from the white-rot fungus P. chrysosporium plays a role in bio-pulping and bio-bleaching (Vandana et al. 2019). Several fungal species producing LiP enzymes, which are believed to play an important role in the biodegradation of lignin, are also known (Le Roes-Hill et al. 2011; Janusz et al. 2017; Vandana et al. 2019).
Lignin peroxidase is also known to be an essential enzyme in wastewater treatment. In principle, it could replace the use of chemical compounds for degrading paper waste; the reduced 5-fold concentration of lignin was observed after adding LiP enzymes in wastewater as compared with untreated wastewater, which is necessary due to the presence of lignin and its derivatives in pulp effluent increasing the pollution in the environment (Gaur et al. 2018). Furthermore, LiP is also known to play a role in the decolorization of dyes in textile industrial wastewater, which was also demonstrated by Parshetti et al. (2012) and Moubasher et al. (2017). It is predicted that the demand for LiP will increase in the future.
Currently, for biological treatments, various fungi show characteristics in the delignification of the lignocellulosic biomass and show potential for dye-decolorization due to their ligninolytic enzymes (Chia 2008; Yanto et al. 2019). Numerous ligninolytic enzymes have been isolated from fungal sources including white-rot fungi, such as Phanerochaete chrysosporium (Vandana et al. 2019), Phanerochaete sordida (Hirai et al. 2005), Trametes versicolor (Jönsson et al. 1994), brown-rot fungi such as Coniophora puteana (Irbe et al. 2014), and Laetiporus sulphureus (Mtui and Masalu 2008). However, among various fungi, the use of white-rot fungi Lentinus squarrosulus has been limited to application in the decolorization of dyes, and its role in the degradation of lignin has not been explored thoroughly.
Recently, Ravichandran et al. (2019) reported a versatile peroxidase from L. squarrosulus and its application in the biodegradation of lignocellulosic materials. However, the secretion of LiP by this white-rot fungus has not been comprehensively elucidated yet. Additionally, the application of this enzyme has not yet been developed properly due to the limited availability; the development of cost-effective production processes as well as purification is still difficult. Hence, in the present study, to gain insight into the redox potentials of LiP from the fungal system, the authors describe the production, purification, and characterization of a LiP from the newly isolated white-rot fungus Lentinus squarrosulus MPN12 (LsLiP) and its use for the bioconversion of lignin and different dyes.
EXPERIMENTAL
Materials and Methods
Fungal isolation and cultural conditions
The fungus (initially designated as MPN12) was collected from deciduous deadwood in the Muong Phang primeval forest (21°27’N, 103°09’E) in Dien Bien Province, Northwest Vietnam. The fungus was grown on a 2% malt-agar plate with antibiotic additions (0.005% streptomycin, penicillin). The seed culture was prepared by repeated sub-culturing on potato dextrose agar (PDA; Merck, Darmstadt, Germany) and stored at –80 °C for further experiments.
Molecular identification and phylogenetic analysis
For fungal identification, the ITS1–5.8S–ITS2 region was amplified from the genome of L. squarrosulus using a polymerase chain reaction (PCR) with a forward primer ITS1: 5ʹ-TCCGTAGGTGAACCTGCGG-3ʹ and a reverse primer ITS4: 5ʹ-TCCTCCGCTTATTGATATTGC-3ʹ (White et al. 1990). For the reaction, 12.5 µL of PCR Master mix kit (2X) (Thermo Fisher, Billerica, MA, USA ), 3 µL DNA (deoxyribonucleic acid), 10 µM of each primer and nuclease-free water up to 25 µL were used. The thermal cycling conditions were 95 °C/3 min followed by 35 cycles of 94 °C/45 s, 55 °C/45 s, and 72 °C/45 s, and a final extension was performed at 72 °C/10 min. The amplification of the products was verified by electrophoresis using a 1.5% agarose gel and purified using QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). Next, amplicons were sequenced in an ABI Prism 3100 genetic analyzer (Applied Biosystems, Waltham, MA, USA). The sequences obtained were analyzed in the ChromasPro1.7.6 software (Technelysium, Brisbane, Australia) and compared with GenBank databases using the NCBI BLAST tool (Johnson et al. 2008).
Phylogenetic analysis
Phylogenetic tree analysis was performed utilizing the MEGA® v6.0 software (Pennsylvania State University, University Park, PA, USA) (Tamura et al. 2013). The alignment was carried out using Clustal W (European Molecular Biology Laboratory, Cambridge, CB10 1SD, United Kingdom) (Thompson et al. 1997), and the evolutionary history was inferred using the maximum likelihood method (ML) (Harris and Stöcker 1998) based on Kimura’s two-parameter model (Kimura 1980) with 1000 bootstrap replicates. TreeView software (S&N Genealogy, Salisbury, United Kingdom) was used to edit evolutionary tree images (Page 1996).
Production and purification of lignin peroxidase
The production of the LsLiP of L. squarrosulus (strain MPN12) was performed in sterile plastic bags, each containing approximately 500 g of rice straw, and trace elements (KH2PO4 0.5 g L-1, MgSO4 0.25 g L-1, (NH4)2SO4 1.5 g L-1, CaCl2 0.05 g L-1, and Na2HPO4.12H2O 2.3 g L-1). After two weeks of fungal growth, the proteins were extracted with distilled water. Then the mycelium and straw particles were removed by centrifugation (10,000 × g for 10 min) and filtration (filter GF6; Whatman PLC, Buckinghamshire, UK). The clear supernatant was harvested and concentrated by a tangential flow ultrafiltration system at 11 °C (10 kDa cut-off; Sartorius, Göttingen, Germany).
The enzyme purification was then performed by three steps of fast protein liquid chromatography (FPLC) using an ÄKTA Pure system (GE Healthcare, Danderyd, Sweden) with a detector operating at 280 nm. Firstly, the protein solution was applied to a HiTrapTM DEAE cellulose column equilibrated with 10 mM sodium acetate buffer (pH 4.5) and using a flow rate of 1 mL/min, eluted using a NaCl gradient of 0 to 1.5 M. LiP-active fractions were pooled and applied to Sephadex G-100 gel filtration column equilibrated with acetate buffer pH 4.5. The final purification step occurred on a HiTrapTM Q XL with 50 mM sodium acetate buffer (pH 5.5) and a NaCl gradient of 0.25 to 1.0 M at a flow rate of 0.5 mL/min. Pooled fractions with LiP activity were concentrated and desalted in the sodium acetate buffer (pH 5.5, 50 mM NaCl) using Amicon® Ultra centrifugal filters (10 kDa cut-off; Sartorius, Göttingen, Germany) and were stored at –80 °C for further use.
Enzyme activity assay
Peroxidase activity was assayed by using 2,4-dichlorophenol (2,4-DCP) (Sigma-Aldrich, Stockholm, Sweden) as the substrate. The reaction mixture (1 mL) contained 100 mM sodium acetate buffer (pH 5.5), 82 mM 4-aminoantipyrine (Sigma-Aldrich, Stockholm, Sweden), 20 mM 2,4-DCP, 40 mM hydrogen peroxide, and an appropriate amount of LsLiP (Yee and Wood 1996). The reaction was initiated by the addition of hydrogen peroxide, during the first 2 min of the reaction at 35 °C at 510 nm. One unit of enzyme activity was defined as the amount of enzyme that transformed 1 µmol of substrate per min.
Enzyme characterization
The weight-averaged molecular weight (Mw) of the purified enzyme was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12%) (Laemmli 1970) at room temperature. The enzyme sample was incubated at 95 °C for 10 min and centrifuged at 2000 rpm for 10 min. Analytical isoelectric focusing (IEF) was performed with the same electrophoresis system but using IEF precast gels (Novex IEF; Invitrogen, Massachusetts, USA). Staining of the protein bands was done using Coomassie Brilliant Blue R-250, which was followed by fixing and destaining.
The effect of pH on LsLiP activity was determined by conducting an enzyme assay within a pH range from 4.0 to 8.0 in 100 mM sodium acetate buffer (pH 4.0 to 5.5) and 100 mM sodium phosphate (pH 6.0 to 8.0). The optimal temperature was determined by measuring the enzyme activity at 20 to 55 °C. The temperature optimum was determined by following the oxidation of 2,4-DCP at different temperatures (20 to 55 °C) in sodium acetate buffer (100 mM) at pH 5.5. The pH stability of LsLiP was tested by incubating the enzyme activity at 35 °C for different time periods in 100 mM sodium acetate buffer, pH 4.0 as well as in sodium phosphate buffer, pH 6.0 and pH 8.0. After 1 h-intervals of incubation, aliquots of the sample were taken, and the remaining LiP activity was measured as described above. The thermal stability of LsLiP was determined by measuring the residual activity after enzyme incubation at 4, 25, 40, and 60 °C at an interval time of 20 min for up to 2 h. Furthermore, various metal ions, e.g., Na+, K+, Ca2+, Cu2+, Zn2+, Mg2+, Ni2+, and Fe2+ at 1 mM and 5 mM were used to elucidate their effects on the activity of LsLiP. All experiments were performed in triplicate.
Substrate specificity and kinetic analysis
Several phenolic compounds were tested as potential LiP substrates, including DCP, veratryl alcohol, guaiacol, ferulic acid, vanillic acid, and dimethoxyphenol (concentration 20 mM). The rate of substrate oxidation was measured by determining the increase in absorbance at their wavelengths.
The Michaelis-Menten constant (Km) and catalytic rate constant (kcat) were determined by measuring the activity of LsLiP at several fixed concentrations of DCP (0.3 to 1.5 mM) at 35 °C for 2 min. The Vmax and Km parameters were calculated by applying the Lineweaver-Burk plot. Data correspond to the value ± standard deviation (SD) of three independent experiments.
Lignin biodegradation
Lignin was extracted from rice-straw biomass, which was first pretreated with ultrasound irradiation (Sonic system SOMERSET) provided with the ultimate power of 500 W under different times (0, 10, 20, 30, and 40 min) in 2 M NaOH aqueous solution (1 g rice straw: 20 ml NaOH 2 M) at 90 °C. Then, the mixture was continuously (150 rpm) stirred at 90 °C for 1.5 h. Next, the mixture was washed with 0.1 M NaOH to separate lignin on the cellulose surface. After the filtration on a nylon cloth, the residue rich in cellulose was removed. The mixture of hydrolysates was then isolated to hemicellulose by precipitation of the acidified hydrolysate (pH was adjusted to 5.5 with HCl solution) with three volumes of 95% ethanol for 6 h. The pellets rich in hemicellulose were filtered and removed. As the last step, after the evaporation of ethanol, the alkali-soluble lignin was obtained by precipitation at pH 1.5 adjusted by HCl. The solid rich in lignin were then washed with acidified solution pH 2.0 and freeze-dried (Dinh et al. 2017).
A total of 10 mL of reaction mixture containing 0.1 g of lignin, 3.3 mL of ethanol (96%), 40 mM H2O2, and 25 U of purified LsLiP in 100 mM sodium acetate at pH 5.0 was incubated at 37 °C for 72 h with shaking at 150 rpm. The changes in lignin biodegradation were identified using Fourier transform infrared spectroscopy (FTIR) analysis, which was determined using a Nicolet iN10 FTIR Microscope (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a DTGS detector.
Table 1. Characteristics of Studied Dyes
Dye decolorization
The decolorizing ability of 6 different dyes belonging to the azo and anthraquinone group by purified LsLiP was determined in the 1 mL-mixtures containing each dye at the final concentration of 0.15 mg/mL, 40 mM H2O2, and 0.25 U of purified LsLiP in 100 mM sodium acetate buffer (pH 5.0). Negative control was measured under the same conditions with the heat-denatured enzyme. After incubation at 25 °C for 12 and 48 h, decolorization was assayed by measuring the absorbance at the respective wavelength maximum of each dye (Table 1). Each decolorization experiment was performed in triplicate, and the mean of the decolorization percentages was reported. The percentage decolorization was determined using Eq. 1,
(1)
where Ai is the initial absorbances of the mixture and At is the final absorbance of the mixture (Vandana et al. 2019).
RESULTS AND DISCUSSION
Identification of Fungal Strains Isolated with High LiP Activity
Identification of the fungal isolates was conducted through partial gene sequencing. Total DNA was extracted and amplified by PCR with primers (ITS1/ITS4). A sequence of 427 nucleotides in length was obtained and deposited in the GenBank nucleotides database (http://www.ncbi.nlm.nih.gov/genbank/) with the accession number MW879106. The sequence obtained was then compared with those available in the GenBank database by applying the NCBI BLAST tool. The results showed 99.76% similarities with the sequences of the Lentinus squarrosulus GU001951 species. Based on this genetic identification, the strain MPN12 was authenticated as Lentinus squarrosulus (Polyporaceae, Basidiomycota), which is a new species for Vietnam. Phylogenetic trees of fungi (ITS region) are shown in Fig. 1; the bootstrap values demonstrate the quality of the analyses.
Purification of LiP from L. squarrosulus
For purification studies, a large amount of LsLiP was produced under conditions of solid-state fermentation with rice straw as a growth substrate to obtain a crude extract with a total LsLiP activity of 3600 U (measured with DCP, present H2O2). Subsequently, LiP from L. squarrosulus was successfully purified using three-step column chromatography procedures including DEAE, Sephadex G-100, and HiTrapTM Q XL columns. At the first step of purification, the specific activity of LsLiP was increased 6.7-fold from 0.6 to 4 U mg-1 (Table 2) after separation by the anion exchanger DEAE-cellulose. Pooled fractions of total activity of 1250 U were subsequently applied to size exclusion chromatography (GE Healthcare, Danderyd, Sweden) with the SephadexTM G-100 column.
Table 2. Purification Summary of LsLiP from L. squarrosulus
Fig. 1. Internal transcribed spacer (ITS) region was used to identify the fungal taxonomy (a), phylogeny tree was performed based on ML method using a partial rDNA-ITS sequence of the MPN12 fungal strain; Psathyrella pygmaea MG734744 was used as an outgroup taxon (b), and white-rot fungus Lentinus squarrosulus MPN12 (c).
These active fractions were further separated using a HiTrapTM Q XL column and a major peak of LiP activity was collected at a NaCl gradient of 0.5 mM (Fig. 2a). The LiP-active protein was purified with a total activity of 566 U with up to 47.1-fold purification and a 15.7% recovery (Table 2).
The purified LsLiP after the final step of HiTrapTM Q XL columns was assessed by 12% SDS-PAGE analysis, which appeared as a single band with a molecular mass of 55 kDa. Purified LsLiP revealed an acidic isoelectric pH value (pI) of 4.5 in the corresponding native IEF gel after staining with colloidal blue (Fig. 2b).
Fig. 2. (a) FPLC elution profile of the first and final purification steps of LsLiP: HiTrapTM Q XL column: absorbance at 280 nm (solid line), LiP activity (black circles), and NaCl gradient (dashed line) and (b) SDS-PAGE gel and IEF analysis of purified LsLiP: (lane 2,3), protein marker (lane 1,4)
Effect of pH and Temperature on the Activity of LsLiP
The optimal pH of LsLiP was determined to be between pH 4.0 and 8.0. The highest activity was observed at pH 5.0 in sodium acetate buffer, and the LsLiP lost its activity at pH 8.0 in sodium phosphate buffer (Fig. 3a). This optimal pH range of LsLiP is similar to those of other LiPs (Alam et al. 2009). The LsLiP exhibited optimal temperatures at 35 °C and approximately 10% of the maximum activity at 55 °C (Fig. 3b).
Fig. 3. Effects of pH and temperature on the activity of LsLiP: (a) Optimum pH: LsLiP was determined at various pH levels (pH 4.0 to 8.0) at 35 °C; (b) Optimum temperature: LsLiP activity was measured at pH 5.0 with different temperatures (20 to 55 °C); (c) pH stability: pH stability of LsLiP was determined by measuring the residual activity at 35 °C after incubation of the enzyme at pH 4, 6, and 8 for the indicated times; (d) Thermal stability: Thermal stability of LsLiP was determined by measuring the residual activity at pH 5.0 after incubation of the enzyme at 4 °C, 25 °C, 40 °C, and 60 °C for the indicated times. Each value represents the mean ± SD of three measurements.
The stability of LsLiP at different pHs was determined by measuring residual activity at optimum conditions (pH 5.0 and 35 °C) after the incubation of the enzyme at various pH (4.0 to 8.0) and for the indicated times. The LsLiP maintained peroxidase activity at pH 4.0 range for 3.0 h and was gradually denatured at pH 6.0. At pH 8.0, LsLiP lost its activity rapidly (Fig. 3c).
The thermal stability of LsLiP was determined by measuring the enzymatic activity at the optimal operating conditions and incubation at various temperatures (4 to 60 °C) for the indicated times. The LsLiP maintained its activity at 4 to 25 °C for 1 h. However, at 40 °C, the activity decreased to 50% and 75% of the initial activity after 1 and 2 h, respectively. The LsLiP activity rapidly reduced at 60 °C (Fig. 3d).
Effect of Metal Ions on the LsLiP Activity
The investigation of the effect of different metal cations (1 and 5 mM) on peroxidase activity of purified LsLiP showed that its activity was slightly enhanced by adding divalent cations, i.e., Cu2+, Fe2+, Ca2+, Zn2+, Mg2+, and Ni2+, at concentrations of both 1 and 5 mM. These results illustrated that divalent cations were required for the activity of LsLiP. This was consistent with previous studies. Louie and Meade (1999) showed that Fe2+ can coordinate with oxidative site residues, leading to the activation of enzymes, whereas Fodil et al. (2012) showed that Ca2+ and Cu2+ were required for the stabilization in the active site of peroxidases. However, the addition of monovalent cation did not affect LsLiP activity (Table 3).
Table 3. Effect of Metal Ions on LsLiP Activity
Substrate Specificity and Kinetic Analysis
Additionally, LsLiP showed a substrate preference for DCP among the phenolic compounds investigated (Fig. 4).
Fig. 4. Effect of the different substrates on the LsLiP activity
It showed up to 240% activity for DCP, which was followed by 180% activity for veratryl alcohol compared with that of the control. However, LsLiP showed much lower activity against other aromatic compounds (guaiacol, ferulic acid, and vanillic acid) (Fig. 4). Broad substrate, specifically, is commonly observed for most of the known LiPs, among which veratryl alcohol is the most frequently preferred (Vandana et al. 2019).
Kinetic constants of purified LsLiP were studied with DCP as substrates. Various concentrations of DCP ranging from 0.3 to 1.5 mM were plotted against the respective initial specific activities (V) of the purified LsLiP. A Lineweaver-Burk reciprocal plot was constructed, and the Michaelis Menten kinetics yielded a hyperbolic curve (Fig. 5). The LsLiP showed a Km value of 2.45 µM and a Vmax value of 285 µmol/min. The catalytic rate (kcat) value of the LsLiP peroxidase reaction was 12 s-1, which led to a catalytic efficiency (kcat/Km) of 4.85 µM-1 s-1.
Fig. 5. (a) Lineweaver-Burk plot and (b) Michaelis Menten plot for the purified LsLiP
Lignin Biodegradation by LsLiP
To determine the biodegradation of lignin by the LsLiP enzyme, the current authors used lignin derived from rice straw as a raw substrate (Dinh et al. 2017). The FTIR spectra of lignin before and after LsLiP treatment are shown in Fig. 6, and the major bands are assigned from a summary compiled from previous studies (Table 4) (Stark et al. 2016; Deng et al. 2019). The fingerprint region (1600 to 1000 cm-1) in the spectra of lignin noticeably changed after using LsLiP treatment, corresponding to the stretching vibrations of various groups in lignin (Ying et al. 2018). This shows that the overall structure of lignin had been destroyed to a certain extent, after enzyme treatment. The stretching vibration of the C-O bonds in aliphatic alcohols and aliphatic ethers-derived aromatic ethers at 1115 cm-1 suggested that the band at 1115 cm-1 was caused by ether linkages adsorption. Next, the intensity of the peak at 1216 cm-1 changed noticeably, suggesting that the carbonyl in guaiacyl was interrupted during the treatment. The intensity of lignin at 1450 cm-1 decreased, indicating that the C-H of the aromatic ring in lignin could be deformed. This remarkable change at 1593 cm-1 was mainly attributed to the stretching of the aryl ring in lignin, and the peak intensity decreased after enzymatic treatment. The appearance of a peak of 1622 cm-1 was related to vibrations of aromatic rings present on lignin, which allowed for the prediction that the conjugated double bonds were generated.
Table 4. Assignments of FTIR Peaks in Lignin Samples
Fig. 6. FTIR spectra of lignin samples before and after LsLiP treatment
The intensity of the peak at 2930 cm-1 decreased noticeably after LiP treatment, which was mainly attributed to the C–H stretching vibration of the methyl and methylene groups in lignin. The band strikingly increased in the region 3200 to 3400 cm-1, which was assigned to O-H stretching vibrations. This band was caused by the presence of alcoholic and phenol hydroxyl groups. Taken together, the FTIR analysis results of the present study indicated that the peak intensity of lignin structure properties remarkably changed after LsLiP treatment, elucidating the potential of LsLiP in the finely-tuned degradation of lignin polymers.
Dye Decolorization by LsLiP
The decolorization of six different dyes by using LsLiP was determined for 12 h and 48 h incubation, at pH 5.0. The results showed that the heat-denatured enzyme used as negative control clearly did not decolorize the tested dyes. Moreover, the decolorization increased with time incubation. In the decolorization of all the dyes at pH 5.0, NY3 was decolorized with higher efficiency than other dyes, while NY1 was difficult to decolorize. The decolorization of dyes was 63%, 50%, 54%, 20.5%, 42.5%, and 41% for NY3, NY5, PB, NY1, IN13, and NY7, respectively, at 12 h incubation. The decolorization level of all dyes remarkably increased to 92%, 70.5%, 73.5%, 34.5%, 61%, and 56% for NY3, NY5, PB, NY1, IN13, and NY7, respectively, at 48 h incubation (Fig. 7a). Moreover, Fig. 7b reveals that the color of the dye was removed after 48 h treatment in the order of NY3 > PB > NY5 > IN3 > NY7 > NY1. The decolorization level of azo dyes was lower than anthraquinone dyes. This could explain why even small structural differences in dyes can be expected to affect the decolorization level of dyes, which has also been investigated by Park et al. (2004).
Fig. 7. Percentage decolorization of dyes by LsLiP
Discussion
The ligninolytic enzymes such as lignin peroxidase that are predominantly secreted by white-rot fungi are varied in the catalytic characteristics with the host species and their ecological recess. Accordingly, the present study reports the characteristics of a LiP purified from white-rot fungi Lentinus squarrosulus, which was isolated in the primeval forest, Vietnam. The enzyme was purified with 47.1-fold purification through anion and size exclusion chromatographic procedures. The purified protein showed a molecular weight of 55 kDa (Fig. 2b), which was slightly higher than LiPs from other fungi previously reported (38 to 46 kDa) (Sahadevan et al. 2016; Chandra and Madakka 2019) but marginally lower than that from bacterial sources (66 kDa) (Ghodake et al. 2009; Parshetti et al. 2012; Patil 2014)
Lignin peroxidase was able to oxidize several phenolic compounds in a wide variety of applications. Among the aromatic compounds, DCP and veratryl alcohol as the main substrate of LiP have been studied the most. However, numerous previous studies have reported the kinetic characteristics of LiP using veratryl alcohol as the substrate (Ollikka et al. 1993; Koduri and Tien 1994; Lee et al. 2001; Casciello et al. 2017; Romero et al. 2019 Vandana et al. 2019), but only a few have reported the kinetics of DCP as the substrate (Yee and Wood 1997; Ryu et al. 2008). In this study, the authors showed a LiP from L. squarrosulus (LsLiP) that catalyzes the oxidation of DCP, veratryl alcohol, and other phenolic compounds. Of the different substrates tested, DCP was the substrate that was subjected to maximum oxidation by LsLiP. Accordingly, the kinetic parameters for the oxidation of DCP by LsLiP in the presence of H2O2 were investigated (Fig. 5). The kinetic data revealed a noticeable difference in the Km value for DCP of LsLiP and other LiPs. Thus, the Km value of LsLiP was 2.45 µM, which was lower than that of a bacterium Streptomyces viridosporus (372 µM) (Yee and Wood 1997), and white-rot fungus P. chrysosporium (359 µM) (Ryu et al. 2008). Moreover, the catalytic efficiency (kcat/Km) of LsLiP (4.85 µM-1 s-1) was higher than that of LiP from white-rot fungus P. chrysosporium (6.34 µM-1 min-1), indicating that LsLiP exhibits a strong affinity against this substrate. The pH profile revealed the optimum pH for DCP oxidation of LsLiP was in the acidic range as for other ligninolytic peroxidases (Couto et al. 2006; Fernández-Fueyo et al. 2014) and its enzymatic activity decreased rapidly beyond the optimal pH (Fig. 3a). The LsLiP maintained its activity at pH 4.0 and gradually lost its activity at pH 6.0 to 8.0 (Fig. 3c). Concerning thermal stability, a temperature higher than 40 °C was not beneficial for the enzyme. The LsLiP exhibited an apparent optimum activity at 35 °C, which was similar to the optimum physiological temperature. At 40 °C, the activity lost approximately 70% of its maximum after incubation for 2 h (Fig. 3d). This could be due to the structural Ca2+ ions of the enzyme that are lost at high temperatures (Ayuso-Fernández et al. 2018).
To ascertain the potential of purified LsLiP, the enzyme was employed to degrade the lignin and decolorize the tested dyes. Fourier-transform infrared spectroscopy analysis showed that lignin structure before and after LsLiP treatment was noticeably changed, especially in region 1600 to 1000 cm-1. Moreover, the intensity of the peak at 2930 cm-1 changed slightly after LsLiP treatment, which was predicted to be the main cause of lignin cleavage. Therefore, LsLiP could degrade lignin in rice straw. This enzyme is similar to other peroxidases, such as versatile peroxidase or manganese peroxidase, which showed the capability to degrade the lignin of the lignocellulosic crop residues (Ravichandran et al. 2019). The LsLiP has also been shown that plays an important role in enhancing the decolorization process of dye as evaluated for six different dyes, including azo and anthraquinone. The optimum incubation time required for dye decoloration remarkably depends on the type and chemical structure of dyes (Sudiana et al. 2018). When the decolorization of different dyes was tested, purified LsLiP showed a higher decolorization rate for anthraquinone dyes than for azo dyes (Fig. 7a). A high decolorization rate was revealed for the anthraquinone dyes, such as NY3 92%, NY5 70.5%, and PB 73.5%, while azo dyes such as NY1, IN13, and NY7 up to 34.5%, 61%, and 56%, within 48 h, respectively. Azo dyes have a complex chemical structure, so they are more difficult to degrade than anthraquinone dyes. Structural bonding in dyes can affect the ability of the LiP enzyme in dye decolorization. Azo dyes have a strong bond between the chromophore group and auxochrome groups, so they are hard to be biologically decolorized by LiP enzymes, whereas anthraquinone dyes have a weak conjugate bond system. When the LiP enzyme attacks the anthraquinone dyes, electrons from the enzyme are transferred to a structure that has conjugate bonds. This transfer changes the configuration of the dye structure and reduces the color intensity. Taken together, the results of the present study indicate that LsLiP from white-rot fungus L. squarrosulus MPN12 is involved in the color removal of synthetic dyes and has potential in the biodegradation of lignin (lignocellulosic biomass). However, for future industrial applications, to improve the degradation efficiency of dyes and lignin, LiP should be combined with other ligninolytic enzymes such as (laccase, manganese peroxidase, versatile peroxidases), which are also high redox potential enzymes from white-rot fungi (Yadav and Yadav 2015).
CONCLUSIONS
- This study is the first to report the secretion of lignin peroxidase (LiP) by the newly isolated Lentinus squarrosulus MPN12 of the white-rot fungus.
- The anion exchange and gel filtration column chromatography techniques were used to purify LiP from L. squarrosulus MPN12 (LsLiP) up to 47.1-fold. The biochemical properties of LsLiP showed that the optimum pH was 5.0, and stability was at the acidic pH range. The optimum temperature was 35 °C for LsLiP and the activity was maintained at a moderate temperature (25 to 30 °C). The purified lignin peroxidase from L. squarrosulus MPN12 (LsLiP) had high specific activity against 2,4-dichlorophenol (DCP) and veratryl alcohol.
- LsLiP from white-rot fungus L. squarrosulus MPN12 is involved in the decolorization of synthetic dyes and has potential for the biodegradation of lignin.
ACKNOWLEDGMENTS
This work is supported by a grant from the Graduate University of Science and Technology, Vietnam Academy of Science and Technology for post-doctoral support (Grant No. GUST.STS.ĐT 2020-HH02) and the Ministry of Science and Technology (NĐT.45.GER/18).
Authors’ Contributions
Vu Dinh Giap and Do Huu Nghi managed the project design and funding acquisition. Do Huu Nghi, Dang Thu Quynh, and Le Huu Cuong managed the experimental performance, data analysis, and manuscript preparation. Vu Dinh Giap and Dang Thu Quynh participated in the data analysis and manuscript revision. Dang Thu Quynh joined in the experimental design for optimization.
REFERENCES CITED
Alam, Z., Mansor, M. F., and Jalal, K. C. A. (2009). “Optimization of lignin peroxidase production and stability by Phanerochaete chrysosporium using sewage-treatment-plant sludge as substrate in a stirred-tank bioreactor,” J. Ind. Microbiol. 36(5), 757-764. DOI: 10.1007/s10295-009-0548-5
Ayuso-Fernández, I., Ruiz-Dueñas, F. J., and Martínez, A. T. (2018). “Evolutionary convergence in lignin-degrading enzymes,” Proc. Natl. Acad. Sci. USA 115(25), 6428-6433. DOI: 10.1073/pnas.1802555115
Casciello, C., Tonin, F., Berini, F., Fasoli, E., Marinelli, F., Pollegioni, L., and Rosini, E. (2017). “A valuable peroxidase activity from the novel species Nonomuraea gerenzanensis growing on alkali lignin,” Biotechnol. Rep. 13, 49-57. DOI: 10.1016/j.btre.2016.12.005
Chandra, M. R. G. S., and Madakka, M. (2019). “Comparative biochemistry and kinetics of microbial lignocellulolytic enzymes,” in: Recent Developments in Applied Microbiology and Biochemistry, V. Buddolla (ed.), Academic Press, Cambridge, MA, USA, pp. 147-159. DOI: 10.1016/B978-0-12-816328-3.00011-8
Chia, C. C. (2008). Enhanced Production of Lignin Peroxidase and Manganese Peroxidase by Phanerochaete chrysosporium in a Submerged Culture Fermentation and Their Application in Decolourisation of Dyes, Thesis, University of Science, George Town, Malaysia.
Christian, V., Shrivastava, R., Shukla, D., Modi, H., and Vyas, B. R. M. (2005). “Mediator role of veratryl alcohol in the lignin peroxidase-catalyzed oxidative decolorization of Remazol Brilliant Blue R,” Enzyme Microb. Technol. 36(2-3), 327-332. DOI: 10.1016/j.enzmictec.2004.09.006
Couto, S. R., Moldes, D., and Sanroman, M. A. (2006). “Optimum stability conditions of pH and temperature for ligninase and manganese-dependent peroxidase from Phanerochaete chrysosporium. Application to in vitro decolorization of Poly R-478 by MnP,” World J. Microbiol. Biotechnol. 22(6), 607-612. DOI: 10.1007/s11274-005-9078-0
Deng, Z., Xia, A., Liao, Q., Zhu, X., Huang, Y., and Fu, Q. (2019). “Laccase pretreatment of wheat straw: Effects of the physicochemical characteristics and the kinetics of enzymatic hydrolysis,” Biotechnol. Biofuels 12, article no. 159. DOI: 10.1186/s13068-019-1499-3
Dinh, V. N., Thi, T. H., Bui, N. D., Duc, V. C., and Viet, N. H. (2017). “Lignin and cellulose extraction from Vietnam’s rice straw using ultrasound-assisted alkaline treatment method,” Int. J. Polym. Sci. (2017), article ID 1063695. DOI: 10.1155/2017/1063695
Fernández-Fueyo, E., Ruiz-Dueñas, F. J., and Martínez, A. T. (2014). “Engineering a fungal peroxidase that degrades lignin at very acidic pH,” Biotechnol. Biofuels 7(1), article no. 114. DOI: 10.1186/1754-6834-7-114
Fodil, D., Jaouadi, B., Badis, A., Nadia, Z. J., Ferradji, F. Z., Bejar, S., and Boutoumi, H. (2012). “A thermostable humic acid peroxidase from Streptomyces sp. strain AH4: Purification and biochemical characterization,” Bioresource Technol. 111, 383-390. DOI: 10.1016/j.biortech.2012.01.153
Garguiak, J. D., and Lebo, S. E. (2000). “Commercial use of lignin-based materials,” in: Lignin: Historical, Biological, and Materials Perspectives, W. G. Glasser, R. A. Northey, and T. S. Schultz (eds.), ACS Publications, Washington D.C., USA, pp. 304-320. DOI: 10.1021/bk-2000-0742.ch015
Gaur, N., Narasimhulu, K., and Setty, Y. P. (2018). “Extraction of ligninolytic enzymes from novel Klebsiella pneumoniae strains and its application in wastewater treatment,” Appl. Water. Sci. 8(4), article no. 111. DOI: 10.1007/s13201-018-0758-y
Ghodake, G. S., Kalme, S. D., Jadhav, J. P., and Govindwar, S. P. (2009). “Purification and partial characterization of lignin peroxidase from Acinetobacter calcoaceticus NCIM 2890 and its application in decolorization of textile dyes,” Appl. Biochem. Biotechnol. 152(1), 6-14. DOI: 10.1007/s12010-008-8258-4
Hamid, M., and Rehman, K. U. (2009). “Potential applications of peroxidases,” Food Chem. 115(4), 1177-1186. DOI: 10.1016/j.foodchem.2009.02.035
Harris, J. W., and Stöcker, H. (1998). “Probability theory and mathematical statistics,” in: Handbook of Mathematics and Computational Science, Springer, New York, NY, USA, pp. 773-844.
Hirai, H., Sugiura, M., Kawai, S., and Nishida, T. (2005). “Characteristics of novel lignin peroxidases produced by white-rot fungus Phanerochaete sordida YK-624,” FEMS Microbiol. Lett. 246(1), 19-24. DOI: 10.1016/j.femsle.2005.03.032
Irbe, I., Elisashvili, V., Asatiani, M. D., Janberga, A., Andersone, I., Andersons, B., Biziks, V., and Grinins, J. (2014). “Lignocellulolytic activity of Coniophora puteana and Trametes versicolor in fermentation of wheat bran and decay of hydrothermally modified hardwoods,” Int. Biodeter. Biodegrad. 86, 71-78. DOI: 10.1016/j.ibiod.2013.06.027
Janusz, G., Pawlik, A., Sulej, J., Świderska-Burek, U., Jarosz-Wilkołazka, A., and Paszczyński, A. (2017). “Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution,” FEMS Microbiol. Rev. 41(6), 941-962. DOI: 10.1093/femsre/fux049
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). “NCBI BLAST: A better web interface,” Nucleic Acids Res. 36(suppl_2), W5-W9. DOI: 10.1093/nar/gkn201
Jönsson, L., Becker, H. G., and Nyman, P. O. (1994). “A novel type of peroxidase gene from the white-rot fungus Trametes versicolor,” Biochim. Biophys. Acta. 1207(2), 255-259. DOI: 10.1016/0167-4838(94)00083-2
Kimura, M. (1980). “A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences,” J. Mol. Evol. 16(2), 110-120. DOI: 10.1007/BF01731581
Koduri, R. S., and Tien, M. (1994). “Kinetic analysis of lignin peroxidase: Explanation for the mediation phenomenon by veratryl alcohol,” Biochemistry. 33(14), 4225-4230. DOI: 10.1021/bi00180a016
Laemmli, U. K. (1970). “Cleavage of structural proteins during the assembly of the head of bacteriophage T4,” Nature 227(5259), 680-685. DOI: 10.1038/227680a0
Le Roes-Hill, M., Rohland, J., and Burton, S. (2011). “Actinobacteria isolated from termite guts as a source of novel oxidative enzymes,” Antonie. Leeuwenhoek 100(4), 589-605. DOI: 10.1007/s10482-011-9614-x
Lee, K. B., Gu, M. B., and Moon, S. (2001). “Kinetics of veratryl alcohol oxidation by lignin peroxidase and in situ generated hydrogen peroxide in an electrochemical reactor,” Eng. Life. Sci. 1(6), 237-245. DOI: 10.1002/1618-2863(200112)1:6<237::AID-ELSC237>3.0.CO;2-Q
Louie, A. Y., and Meade, T. J. (1999). “Metal complexes as enzyme inhibitors,” Chemical Rev. 99(9), 2711-2734. DOI: 10.1021/cr9804285
Moubasher, H., Mostafa, F. A., Wahsh, S., and Haroun, O. (2017). “Purification and characterization of lignin peroxidase isozymes from Humicola grisea (Traaen) and its application in bioremediation of textile dyes,” Egypt. J. Bot. 57(2), 335-343. DOI: 10.21608/EJBO.2017.775.1045
Mtui, G., and Masalu, R. (2008). “Extracellular enzymes from brown-rot fungus Laetiporus sulphureus isolated from mangrove forests of coastal Tanzania,” Sci. Res. Essays 3(4), 154-161.
Ollikka, P., Alhonmäki, K., Leppänen, V., Glumoff, T., Raijola, T., and Suominen, I. (1993). “Decolorization of azo, triphenyl methane, heterocyclic, and polymeric dyes by lignin peroxidase isoenzymes from Phanerochaete chrysosporium,” Appl. Environ. Microbiol. 59(12), 4010-4016. DOI: 10.1128/aem.59.12.4010-4016.1993
Page, R. D. (1996). “Tree view: An application to display phylogenetic trees on personal computers,” Bioinformatics 12(4), 357-358. DOI: 10.1093/bioinformatics/12.4.357
Pandit, S., Savla, N. R., Sonawane, J. M., Sani, A. M., Gupta, P. K., Mathuriya, A. S., Rai, A. K., Jadhav, D. A., Jung, S. P., and Prasad, R. (2021). “Agricultural waste and wastewater as feedstock for bioelectricity generation using microbial fuel cells: Recent advances,” Fermentation 7(3), article no. 169. DOI: 10.3390/fermentation7030169
Park, C., Lee, Y., Kim, T., Lee, B., Lee, J., and Kim, S. (2004). “Decolorization of three acid dyes by enzymes from fungal strains,” J. Microbiol. Biotechnol. 14(6), 1190-1195.
Parshetti, G. K., Parshetti, S., Kalyani, D. C., Doong, R., and Govindwar, S. P. (2012). “Industrial dye decolorizing lignin peroxidase from Kocuria rosea MTCC 1532,” Ann. Microbiol. 62(1), 217-223. DOI: 10.1007/s13213-011-0249-y
Patil, S. R. (2014). “Production and purification of lignin peroxidase from Bacillus megaterium and its application in bioremediation,” CIBTech J. Microbiol. 3, 22-28.
Ravichandran, A., Rao, R. G., Gopinath, M., and Sridhar, M. (2019). “Purification and characterization of versatile peroxidase from Lentinus squarrosulus and its application in biodegradation of lignocellulosics,” J. Appl. Biotechnol. Bioeng. 6(6), 280-286. DOI: 10.15406/jabb.2019.06.00205
Romero, J. O., Fernández-Fueyo, E., Avila-Salas, F., Recabarren, R., Alzate-Morales, J., and Martínez, A. T. (2019). “Binding and catalytic mechanisms of veratryl alcohol oxidation by lignin peroxidase: A theoretical and experimental study,” Comput. Struct. Biotechnol. J. 17, 1066-1074. DOI: 10.1016/j.csbj.2019.07.002
Ryu, K., Kang, J. H., Wang, L., and Lee, E. K. (2008). “Expression in yeast of secreted lignin peroxidase with improved 2,4-dichlorophenol degradability by DNA shuffling,” J. Biotechnol. 135(3), 241-246. DOI: 10.1016/j.jbiotec.2008.04.007
Sahadevan, L. D. M., Misra, C. S., and Thankamani, V. (2016). “Characterization of lignin-degrading enzymes (LDEs) from a dimorphic novel fungus and identification of products of enzymatic breakdown of lignin,” 3 Biotech. 6(1), article no. 56. DOI: 10.1007/s13205-016-0384-z
Stark, N. M., Yelle, D. J., and Agarwal, U. P. (2016). “Techniques for characterizing lignin,” in: Lignin in Polymer Composites, O. Faruk, and M. Sain (eds.), Elsevier, Amsterdam, Netherlands, pp. 49-66. DOI: 10.1016/B978-0-323-35565-0.00004-7
Sudiana, I. K., Sastrawidana, I. D. K., and Sukarta, I. N. (2018). “Decolorization study of remazol black B textile dye using local fungi of Ganoderma sp. and their ligninolytic enzymes,” Environ. Sci. Technol. 11(1), 16-22. DOI: 10.3923/jest.2018.16.22
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). “MEGA6: Molecular evolutionary genetics analysis version 6.0,” Mol. Biol. 30(12), 2725-2729. DOI: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). “The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools,” Nucleic Acids Res. 25(24), 4876-4882. DOI: 10.1093/nar/25.24.4876.
Tien, M., and Kirk, T. K. (1983). “Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium Burds,” Science 221(4611), 661-663. DOI: 10.1126/science.221.4611.661
Tien, M., and Kirk, T. K. (1984). “Lignin-degrading enzyme from Phanerochaete chrysosporium: Purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase,” Proc. Natl. Acad. Sci. USA 81(8), 2280-2284. DOI: 10.1073/pnas.81.8.2280
Vandana, T., Kumar, S. A., Swaraj, S., and Manpal, S. (2019). “Purification, characterization, and biodelignification potential of lignin peroxidase from immobilized Phanerochaete chrysosporium,” BioResources 14(3), 5380-5399. DOI: 10.15376/biores.14.3.5380-5399
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in: PCR Protocols: A Guide to Methods and Applications, M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (eds.), Academic Press, Cambridge, MA, USA, pp. 315-322. DOI: 10.1016/B978-0-12-372180-8.50042-1
Yadav, M., and Yadav, H. S. (2015). “Applications of ligninolytic enzymes to pollutants, wastewater, dyes, soil, coal, paper and polymers,” Environ. Chem. Lett.. 13(3), 309-318. DOI: 10.1007/s10311-015-0516-4
Yanto, D. H. Y., Auliana, N., Anita, S. H., and Watanabe, T. (2019). “Decolorization of synthetic textile dyes by laccase from newly isolated Trametes hirsuta EDN084 mediated by violuric acid,” Earth Environ. Sci. 374, Article ID 012005. DOI: 10.1088/1755-1315/374/1/012005
Yee, D. C., and Wood, T. K. (1996). “Elicitation of lignin peroxidase in Streptomyces lividans,” Appl. Biochem. Biotechnol. 60(2), 139-149. DOI: 10.1007/BF02788068
Yee, D. C., and Wood, T. K. (1997). “2,4‐Dichlorophenol degradation using Streptomyces viridosporus T7A lignin peroxidase,” Biotechnol. Prog. 13(1), 53-59. DOI: 10.1021/bp960091x
Ying, W., Shi, Z., Yang, H., Xu, G., Zheng, Z., and Yang, J. (2018). “Effect of alkaline lignin modification on cellulase–lignin interactions and enzymatic saccharification yield,” Biotechnol. Biofuels 11(1), article no. 214. DOI: 10.1186/s13068-018-1217-6
Zhang, Y., and Naebe, M. (2021). “Lignin: A review on structure, properties, and applications as a light-colored UV absorber,” ACS Sustain. Chem. Eng. 9(4), 1427-1442. DOI: 10.1021/acssuschemeng.0c06998
Article submitted: January 26, 2022; Peer review completed: April 25, 2022; Revised version received and accepted: May 30, 2022; Published: June 9, 2022.
DOI: 10.15376/biores.17.3.4480-4498
