Abstract
Landfill leachate is a serious contaminant for groundwater and surface water because of its potentially toxic metal content. In many countries, leachate is discharged into the natural environment without treatment because of the high disposal cost. However, this environmental problem can be solved by microorganisms, as they can adsorb the contaminants or convert them into end products, and this is cost-effective. This study focused on determining bacteria capable of efficiently removing toxic metals from leachates. Therefore, bacteria were isolated from nature that have a high adsorption and resistance capacity to a number of toxic metals. This potential was achieved by Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum, among others. Their efficiency in removing toxic metals compared to raw leachate was Cd (78%, 67%, 78%), Ni (64%, 57%, 56%), Pb (99%, 75%, 76%), Cr (41%, 46%,19%), Co (45%, 60%, 40%), and Cu (80%, 80%, 60%), respectively. According to the results, these bacterial strains proved to be very effective in the treatment of toxic metals from leachate. Therefore, they are good candidates for the treatment of wastewater by bioremedial methods.
Download PDF
Full Article
Removal Efficacy of Toxic Metals in Leachate through Micro-Organisms Isolated from the Natural Environment
Ayhan Kocaman,a,* Burak Feyyaz Savaş,b and Derya İşler Ceyhan c
Landfill leachate is a serious contaminant for groundwater and surface water because of its potentially toxic metal content. In many countries, leachate is discharged into the natural environment without treatment because of the high disposal cost. However, this environmental problem can be solved by microorganisms, as they can adsorb the contaminants or convert them into end products, and this is cost-effective. This study focused on determining bacteria capable of efficiently removing toxic metals from leachates. Therefore, bacteria were isolated from nature that have a high adsorption and resistance capacity to a number of toxic metals. This potential was achieved by Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum, among others. Their efficiency in removing toxic metals compared to raw leachate was Cd (78%, 67%, 78%), Ni (64%, 57%, 56%), Pb (99%, 75%, 76%), Cr (41%, 46%,19%), Co (45%, 60%, 40%), and Cu (80%, 80%, 60%), respectively. According to the results, these bacterial strains proved to be very effective in the treatment of toxic metals from leachate. Therefore, they are good candidates for the treatment of wastewater by bioremedial methods.
DOI: 10.15376/biores.18.3.5476-5493
Keywords: Bacteria; Leachate; Heavy metal; Water treatment; Wastewater
Contact information: a: Karabük University, Engineering Faculty, Environmental Engineering Department, 78050, Karabük, Turkey; b: Karabük University, Engineering Faculty, Environmental Engineering Department, 78050, Karabük, Turkey; c: Gaziantep University, Faculty of Arts and Science, Department of Biology, Gaziantep, Turkey;
*Corresponding author: ayhan.kocaman.ak@gmail.com
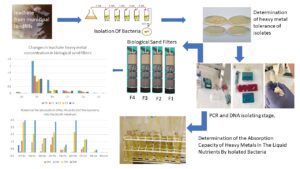
GRAPHICAL ABSTRACT

Pollution of surface and groundwater by potentially toxic metals (PTMs) is a global problem. The PTMs pose chronic and even acute risks to the health of all living organisms, including humans (Imron et al. 2021). One of the sources of concern for human health and the environment is municipal solid waste landfills, which are a major source of soil and groundwater contamination in many regions of the world (Vodyanitskii 2016). Landfill leachate is also becoming increasingly problematic because of its potential for severe groundwater contamination (Vodyanitskii 2016). This is because many contaminants are highly toxic and dangerous to all forms of life. These include heavy metals, such as arsenic (As), chromium (Cr), cadmium (Cd), mercury (Hg), and lead (Pb), which are toxic even at very low concentrations (Jayanthi et al. 2016; Rahman and Singh 2019). These PTMs have been identified as a major threat by international organizations such as the U.S. Environmental Protection Agency (EPA), the World Health Organization (WHO), the Agency for Toxic Substances and the Register of Diseases (ATSDR), and the United Nations Environment Program (UNEP) (ATSDR 2007; Rahman and Singh 2019; Valdiviezo et al. 2021). Leaching of these PTMs from MSW landfills contaminates soil and groundwater, especially drinking water wells. The leachate volume of large landfills with a waste mass of several million tons can exceed 45,000 m3/year or more than 123 m3 per day (Masoner et al. 2016). It has been reported that PTMs leached from landfill leachate at a rate of 400 mm per year can persist in groundwater and surface water for about 150 years (Adelopo et al. 2018). Therefore, effective and cost-efficient leachate treatment technologies are needed for landfill management (Iravanian and Ravari 2020; Ozbay et al. 2021). Conventional treatment methods, although effective, are environmentally harmful to landfill leachate treatment and are also technologically expensive (Dursun 2006; Fan et al. 2008; Hermosilla et al. 2009). Although many chemical and physical methods have been used to treat leachate (Akgul et al. 2013; Gotvajn and Pavko 2015), many authors have confirmed, for example, the ability of carbon as an adsorbent to remove various inorganic and organic pollutants (Al-Saadi et al. 2013; Saleh and Gupta 2014). The use of adsorbents is a widely accepted approach for removing pollutants from wastewater. However, there is still much to be done to improve their efficiency, cost-effectiveness, and ecological performance (Gupta 1998; Saleh 2015a,b). To reduce the harmful effects of landfill leachate, many researchers are still conducting various experiments to treat the leachate effectively.
One of the most effective methods for treating leachate contamination is microbial treatment. One of these methods, bioremediation, is one of the most effective biotechnologies for removing PTMs. Recently, researchers have found that many microbes that break down waste into smaller forms are also among the most promising technologies due to their efficiency, relative cost-effectiveness, and environmental friendliness (Saetang and Babel 2012; Razarinah et al. 2014; Vijayaraghavan and Balasubramanian 2015), and these studies have identified the use of microbial metabolism as an essential part of biotechnology. The mechanism of PTM removal by microorganisms is a rather complex process. Some microorganisms have the potential to produce enzymes (Awasthi et al. 2017), surface binding (Sağ and Kutsal 1996; Tunali and Akar 2006), the mechanism of biosorption of metals, chelation, ion exchange, adsorption (Volesky 2007; Vijayaraghavan and Yun 2008; Wang and Chen 2009; Abdolali et al. 2014). In addition, microprecipitation of metals from outside the cell membrane to intracellular accumulation (Ahalya et al. 2003) suggests that there are related processes for the removal of heavy metals from contaminated areas. Bacteria, such as Pseudomonas sp., Bacillus sp., Acinetobacter sp., Stenotro-phomonas sp., Rhodococcus sp., and Burkholderia sp., have been isolated from actively stored soil and leachate in the literature (Imron et al. 2019). It is also reported that P. aeruginosa isolated from leachate of solid waste landfills can remove about 29, 30, 35, and 28% of Pb, Cr, Ni, and Cd, respectively, and P. aeruginosa KHY2 and Klebsiella pneumonia KHY3 isolated from gold mine wastewater can reduce 1000 mg L-1 Hg by up to 60% (Neneng and Gunawan 2018).
Little is known about the isolation of bacteria from the leachate of inactive landfills (Imron et al. 2021). Leaching metals in inactive landfills occurs not only by physicochemical leaching but also through biological leaching process that produces bacteria that are highly resistant to PTMs (Imron et al. 2021). In addition, bacteria isolated in contaminated areas can remove PTMs and are very resistant. Thus, indigenous microorganisms isolated in contaminated environments can resist and remove contaminants. One study reported that the combination of E. hormaechei and Klebsiella sp. is very effective in removing nitrate, even in the presence of heavy metals. These two bacterial strains exhibit a remarkable ability to tolerate heavy metals, outperforming other denitrifiers. They have shown nitrate removal efficiencies above 99% and higher tolerance to Cu and Zn than many other denitrification strains. Thus, E. hormaechei and Klebsiella sp. are among the heaviest metal-tolerant denitrifying bacterial strains known to date (Amoako-Nimako et al. 2022). In addition, isolated strains of Enterobacter hormaechei have been reported to have significant potential for bioremediation in areas contaminated with nickel and various potentially toxic metals (Heidari et al. 2020), and the bioremediation potential of E. hormaechei and mutagen-exposed E. hormaechei (contaminated with Cd 2+) was investigated in a one-week experiment. It is reported that mutant E. hormaechei exhibited excellent bioremediation potential (90.2%) compared to the wild strain (82.5%) on the sixth day of treatment. Moreover, the significant decrease of Cd2+ concentration in bioremediation started on the third day of treatment and decreased significantly until the sixth day of treatment (Lu et al. 2023). Recently, P. aryabhattai was reported to reduce the phytotoxicity of arsenic in plants and is the most promising PGPR showing great potential for new plant production strategies as it possesses several important PGP properties (Ghosh et al. 2018). Priestia aryabhattai has been shown to be able to degrade benzoate, methyl parathion, and polyethylene terephthalate (Dhaka et al. 2022; Esikova et al. 2021; Le et al. 2021). In addition, a specific strain of Priestia aryabhattai, namely KX-3, isolated from East Antarctica, was reported to be able to remove nitrogen under alkaline pH and low-temperature conditions (Kang et al. 2023). However, there are not many studies on the removal of potentially toxic metals from water by this bacterium. The Mycobacterium group includes pathogens that appear to be a source of infection to humans in the environment (Sanders and Wolinsky 1980), and their abundance is particularly noted in waters with heavy metal pollution (Falkinham et al. 1984; Davis-Hoover 1990), and they have been reported to have the ability to degrade methoxychloroethane, an organochlorine insecticide (Satsuma and Masuda 2012). In addition, effective results have been found in the removal of hydrocarbons, aromatics, and polycyclic hydrocarbons (Park et al. 1998). In conclusion, the diversity of mycobacteria, particularly prokaryotes, has been shown to offer the potential for adaptation to a variety of habitats, including environments heavily contaminated with hydrocarbons and heavy metals. Although mycobacteria are abundant in the environment, they have been overlooked for such an important use, a potential that is unexplored and unapplied in the bioremediation of hazardous chemicals (Azadi et al. 2017). This broad adaptability is also of great value for the bioremediation of degraded ecosystems.
This research aimed to determine which bacterial species have developed tolerance to heavy metals and adapted to natural conditions to obtain them for the National Center for Biotechnology gene bank. In addition, the removal of PTMs from leachate was investigated by producing low-cost and effective biological sand filters from these isolated bacteria. In addition, the removal efficiency of Cd, Pb, and Ni was to be determined under laboratory conditions.
EXPERIMENTAL
The Collection of Leachate and Soil Samples for the Isolation of Bacteria
Samples were collected from leachate and surrounding soils in the area where municipal waste and iron and steel industry waste were stored (41°10’40.20” N and 32°39 9.55” E) in Karabük province. The samples were transferred to sterile bags and brought to the laboratory on ice within 6 h. For isolation of bacterial species, 10 g of the soil samples were weighed and placed in test tubes containing 90 mL of isotonic water and then shaken for 15 min. After shaking, 10 mL of the soil solutions were taken, and serial dilutions were made between 10-1 and 10-5. A total of 0.1 mL of the diluted solutions from 10-3 were placed on a culture medium (NA) and incubated at 37 °C for 48 h. At the end of the 48-h, samples were taken from different growing colonies to obtain pure cultures. All these procedures were also repeated for the leachate samples from the landfill. The pure cultures obtained were stored in a glycerol solution at -20 °C for research.
Determination of Potentially Toxic Metal Tolerance of Bacterial Isolates
The isolated 12 bacterial strains were inoculated in a culture medium containing solutions of 100, 200, 400, 550, and 600 ppm Pb (PbCl), Ni (NiCl2.6H2O), and 0.5, 1, 2, 3, 4, and 5 ppm Cd (CdCl2– 2.5H2O) and then incubated at 37 °C for 48 h. This procedure was performed in three replicates in Petri dishes. At the end of the 48-h incubation period, the bacteria with the highest PTM concentrations were detected in a total of 576 Petri dishes. Three bacterial isolates that exhibited the highest heavy metal tolerance to Cd, Ni, and Pb were selected, and the species were identified and used for the experimental studies in this study.
16S rDNA Gene Region Polymerase Chain Reaction of Bacterial Isolates
To isolate DNA, bacterial isolates were incubated in the new medium at 30 °C for 24 h. After the incubation period, DNA isolation was carried out from each isolated strain, following the instructions given in the Invitrogen DNA isolation kit. 16S RDNA primers were used to amplify 16S RDNA regions from the obtained genomic DNA (27F5′-AGAGTTTGATCCTGGCTCAG and 1492R5′-GGTTACCTTGTTACGACTT) (Bal 2012). The total volume of the PCR response was set at 30 µL. Accordingly, 3 µL of 10x Buffer, 25 mM MgCl2 (3 µL), 2 mM dNTP (3 µL), 10 pmol forward Primer (1.5 µL), 10 pmol reverse Primer (1.5 µL), 1.2 µL Tag DNA polymerase, and 2 µL DNA were used. The PCR conditions were performed with 35 cycles at 94 °C for 30 s, at 55 °C for 30 s, at 72 °C for 1.5 min, with an initial denaturation at 94 ºC for 3 min, and a final elongation at 72 °C for 10 min. After imaging the gene region with a length of approximately 1400 bp obtained through PCR in the Agarose gel electrophoresis, sequences were performed in bidirectional sequence analysis. Sequential procedures were performed at Gaziantep University, Department of Molecular Biology.
Molecular Characterization of Bacterial Isolates Using 16S RDNA Gene Area
To determine the molecular species of the isolates obtained in the study areas, long bands of approximately 1000 to 1400 bps were obtained from the 16S RDNA gene region and analyzed. The sequence results obtained with the front and rear caps were evaluated in the FinchTV graphical viewer program (Geospiza Inc., Finch TV Version 1.4.0, Seattle WA, USA). The results from the sequence were recorded in the Seqtrace.exe program, resulting in a single sequence in FASTA format. The results in FASTA format were compared with the nucleotide sequences of bacterial strains recorded in the GenBank using the National Center for Biotechnology Information (NCBI) nucleotide BLAST program. Molecular identification of isolates was detected under MEGA7 (Kumar et al. 2016). A phylogenetic analysis was performed. The evolutionary history was established by bootstrap consensus (1000 replicates) using the statistical method Joining (Felsenstein 1985; Saitou and Nei 1987). Evolutionary distances were determined using the p-distance method (Nei and Kumar 2000). Clostridium butyricum (EU621841.1) was autographed in the phylogenetic tree.
Biological Sand Filter Preparation
The biological sand filters were made of a 50-cm-long and 20-cm-wide polyvinyl chloride (PVC) pipe. The experiment was performed with 3 replicates for each bacterial strain. The soil and all other materials used for the filter were autoclaved at 150 °C for 2 h. Then, they were added to the tube as follows: I) It was filled with quartz sand filter (1.5 to 3 mm) at the height of 15 cm. II) Pebbles (5 to 12 mm) were placed 10 cm high. III) Larger pebbles (18 to 20 mm) were used at a height of 10 cm. IV) Soil was filled at a height of 15 cm. For the inoculation of soil bacteria, soil samples were mixed in using a sterile blender. On the first and second days, 50 mL of water was sprayed with a microbial load of 10-3. The samples were incubated for a total of 48 h. After 48 h of incubation, the soil samples were placed on the filter systems. The leachate began to drain uniformly from the top, one drop in 10 s. After 24 h, water samples were collected from the bottom of the sand filters and analyzed for heavy metals.
F0: Original raw leachate.
F1: Only sterile soil and filter material was used; no bacterial strain was inoculated.
F2: Soil inoculated with Enterobacter hormaechei strains was used.
F3: Soil inoculated with Priestia aryabhattai strains was used.
F4: Soil inoculated with Mycobacterium sacrum strains was used.
Determination of the Absorption Capacity of Bacterial Strains in a Liquid Nutrient Medium Contaminated with Potentially Toxic Metals
Liquid media containing 10 ppm each of Ni, Pb, and Cd in 100 mL were prepared and their pH was adjusted to 6.5. Then they were poured into sterilized 50 mL incubation cups. Three isolated bacterial strains were inoculated into these sterilized incubation cups in three replicates (108 incubation cups in total) and incubated at 37 °C. After every 24 h (24, 48, 72, and 96 h), the inoculated three different bacterial broths were collected in three replicates and centrifuged at 3000 rpm for 10 min. Three types of leachates have been defined depending on the age of the landfill. Landfill leachate is specified between 6.5 and 7.5 in an age range of 5 to 10 years (Chian and DeWalle 1976). The original pH of the raw leachate from the sampling area was measured to be 6.8, which is consistent with what is reported in the literature. Therefore, the pH of the nutrient broth to which potentially toxic metals were added was adjusted to 6.5.
Toxic Metal Concentration Analysis of Leachate and Bacteria
Leachate passed through filters or raw leachate: Leachate samples were prepared for analysis by acid micro digestion in three steps. (Speed wave/MWS-2 Berghof products + Instruments Harreststr, Enien, Germany) (Mertens 2005). After sample preparation for analysis, the inductively coupled plasma optical emission spectroscope (ICP OES) (Perkinelmer, Optima 2100 DV, Shelton, CT, USA) was used to determine the PTM content. The same procedure was used for the centrifuged bacterial pellets.
Statistical Analysis
All data were analyzed using analysis of variance (ANOVA) SPSS 22 software (IBM, Armonk, NY, USA). Duncan’s test was performed to detect significant differences in means (p < 0.05).
RESULTS
After molecular characterization of the bacterial isolates with the 16S RDNA gene region, they were compared and matched with the nucleotide sequences in the registered GenBank. The bacterial strains were assigned to the same group (Priestia aryabhattai (OP641851), Mycobacterium sacrum (OP641851), Enterobacter hormaechei (OP641850)) in the comparative phylogenetic tree with the reference strains found in NCBI (Fig. 1a, b). In addition, the strains were recorded using the Nucleotide BLAST program in NCBI (National Center for Biotechnology Information).
Fig. 1a. Phylogenetic tree based on 16S rDNA gene sequences
Fig. 1b. Agarose gel electrophoresis image
Statistically significant differences in heavy metal concentrations of leachate were found between F0, F1, and biological sand filters F2, F3, and F4 (p < 0.05) (Table 1).
Table 1. Changes in Heavy Metal Concentration in Leachate in Biological Sand Filters Inoculated with Different Bacteria and Without Bacteria
Table 2. Potential for Absorption of Ni, Pb, and Cd of the Bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum into the Broth Medium
The analyzed PTM concentrations of F0, F1, F2, F3, and F4 were determined: Cd (0.09 ± 0.01, 0.08 ± 0.01, 0.02 ± 0.01, 0.03 ± 0.01 and 0.02 ± 0.01), Ni (2.36 ± 0.01, 1.28 ± 0.01, 0.86 ± 0.01, 1.01 ± 0.00 and1.05 ± 0.01), Pb (0.97 ± 0.01, 0.17 ± 0.01, 0.01 ± 0.00, 0.25 ± 0.00 and 0.23 ± 0.01), Cr (0.22 ± 0.01, 0.19 ± 0.01, 0.13 ± 0.01, 0.12 ± 0.00 and 0.18 ± 0.01), Co (0.20 ± 0.01, 0.16 ± 0.01, 0.11 ± 0.01, 0.08 ± 0.00 and 0.12 ± 0.01), and Cu (0.05 ± 0.01, 0.02 ± 0.01, 0.01 ± 0.01, 0.01 ± 0.00 and 0.02 ± 0.01) ppm, respectively.
Statistically significant differences were found between the absorption capacities of the isolated bacteria into the broth medium containing 10 ppm of Ni, Cd, and Pb and the removal efficiency over time (p < 0.05) (Table 2).
The concentrations of Ni absorbed by the bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum were determined by time series as follows: 24th h (0.10 ± 0.02, 0.01 ± 0.00, and 0.13 ± 0.01), 48th h (1.59 ± 0.01, 1.41 ± 0.01, and 1.24 ± 0.01), 72nd h (4.24 ± 0.01, 6.03 ± 0.01, and 2.00 ± 0.05), 96th h (4.58 ± 0.01, 2.58 ± 0.01, and 1.98 ± 0.01) as ppm (Fig. 2).
Fig. 2. Absorption capacities based on time series of bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum in media containing 10 ppm Ni
Fig. 3. Absorption capacities based on time series of bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum in media containing 10 ppm Pb
The concentrations of Pb absorbed by the bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum were determined by time series as follows: 24th h (1.09 ± .01, 37 ± .05, and 06 ± .01), 48th h (2.16 ± .01, 0.38 ± 0.06, and 0.08 ± 0.01), 72nd h (2.77 ± 0.01, 0.75 ± 0.04, and 2.86 ± 0.04), 96th h (4.29 ± 0.01, 1.09 ± 0.01, and 5.64 ± 0.04) ppm (Fig. 3).
The concentrations of Cd absorbed by the bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum were determined by time series as follows: 24th h (0.45 ± 0.01, 0.01 ± 0.00, and 0.01 ± 0.,00), 48th h (0.76 ± 0.05, 0.02 ± 0.01, and 0.02 ± 0.01), 72nd h (1.76 ± 0.05, 0.25 ± 0.01, and 0.04 ± 0.01), and 96th h (1.83 ± 0.03, 0.36 ± 0.06, and 0.05 ± 0.01) ppm (Fig. 4).
Fig. 4. Absorption capacities based on time series of bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum in media containing 10 ppm Cd
DISCUSSION
As an environmentally friendly technique, bioremediation has excellent potential for treating PTM in contaminated soils and waters. Moreover, microorganisms tolerant to toxic metals are essential for the bioremediation of contaminated areas with high specific levels of toxic metals (Yetunde Mutiat et al. 2018). Bacterial bioremediation can ameliorate contaminants in the environment through various mechanisms, such as adsorption, absorption, sequestration, detoxification, and conversion, to a bioavailable or less toxic and more soluble form (Zhang et al. 2020). This study observed a significant reduction in PTM concentration by F1, F2, F3, and F4 compared to the raw leachate (F0). The treatment rate of PTMs in biofilters was determined as follows: Cd (11%, 78%, 67%, and 78%), Ni (46%, 64%, 57%, and 56%), Pb (83%, 99%, 75%, and 76%), Cr (14%, 41%, 46%, and 19%), Co (20%, 45%, 60%, and 40%), Cu (60%, 80%, 80%, and 60%) F1, F2, F3, and F4, respectively. It was found that the bacterium Enterobacter hormaechei isolated from leachate was more effective than other bacteria in removing PTMs from leachate. The fact that this bacterium isolated from its natural environment was more effective than other bacteria in removing PTMs suggests that it can be used to treat PTMs in leachate, especially wastewater. According to the analysis results of a study comparing the most commonly used systems for wastewater treatment in developing countries, the high-rate trickling filter systems are 0.3 to 0.45 m2/person, 0.5 to 1 W/person, and the construction cost is 40 to 70 US dollars/person. The cost of subsurface and rapid infiltration systems is reported to be 1 to 6 m2/person, 0 W/person, and construction cost is 5 to 15 US dollars /person. It has also been mentioned that conventional wastewater treatment systems have more disadvantages than subsurface and rapid infiltration systems, such as high energy consumption, sensitivity to toxic loads, needs for sophisticated operation, climate conditions, and high construction and operation costs (von Sperling 1996). An effective bacterial-biological sand filtration system at sites where leachate is generated will reduce these costs and provide an alternative solution to a significant environmental and health problem in developing countries. In addition, developing countries are generally located in tropical and subtropical regions. We assume that the microorganisms to be used in biological sand filters are hardly affected by temperature and climatic conditions since they usually operate below the soil surface. We can consider this as an advantage of this system. Furthermore, a constant supply of nutrients and substrate is not necessary due to the leachate content of the landfill. The disadvantage of this system is that it takes up more space, but if we take into account the surplus of unused space around the landfill, this will not prove to be a disadvantage. In addition, the results of the study showed that it is necessary to install a bacterial-biological sand water filter at a low cost in developing countries in areas where municipal waste is dumped. With the biological sand water filters to be installed, the pollutants in the leachate will prevent the pollution of surface water and groundwater and reduce the risk to public health. Some studies suggest that this bacterium should also be used to treat other pollutants, especially in wastewater. One study suggests that E. hormaechei JH can be used for organic deodorization and, along with other bacteria, has the potential to be effective in biological deodorization systems (Kim et al. 2008). In a study conducted in nitrate-polluted waters, Enterobacter hormaechei showed high removal potential with a denitrification rate of 3.0 C/N (98.7%) compared to methanol in the presence of sodium succinate. The results show that it can be accepted as a strong candidate for bio-denitrification of wastewater (Kebabi et al. 2018). Other studies show that the artificially induced mutation in E. coli has excellent tolerance to Cd metal and effectively absorbs Cd within the short duration of the bioremediation process (Kaur and Roy 2021), and Priestia aryabhattai is reported to be effective in the treatment of soil and water contaminated with organophosphates (Le et al. 2021). Mycobacterium is a genus of Actinomycetes that belongs to its own family, Mycobacteriaceae. There are more than 190 known species within the genus (King et al. 2017). The genus includes pathogens that can cause serious diseases in mammals, including tuberculosis in humans (Mycobacterium tuberculosis) and leprosy (Mycobacterium leprae) (Hussain 2007). For this reason, the use of this microorganism in soil and water purification is not recommended because it may pose serious risks to human health and life. However, it is expected that this species will be helpful for future studies if mutations are identified that do not pose a health risk.
After the 24th, 48th, 72nd, and 96th h, the concentrations of Ni, Cd, and Pb absorbed by the bacteria Enterobacter hormaechei, Priestia aryabhattai, and Mycobacterium sacrum were determined as follows: Ni (1%, 15.9%, 42.4%, and 45.8%), (0.1%, 14.1%, 25.3%, and 60.3%), (1.3%, 12.4%, 19.8%, and 20%); Cd (4.5%, 7.6%, 17.6%, and 18.3%), (0.1%, 0.2%, 2.5%, and 3.6%), (0.1%, 0.2%, 0.4%, and 0.5%), and Pb (10.9%, 21.6%, 27.7%, and 42.9%), (3.8%, 3.8%, 7.5%, and 10.9%), (0.6%, 0.8%, 28.6%, and 56.4%). In the study conducted in liquid media, Enterobacter hormaechei and Priestia aryabhattai showed higher Ni, Cd, and Pb absorption potential than Mycobacterium sacrum. However, these three isolated bacterial strains did not show as high Cd absorption capacity as Ni and Pb. This might be due to the stronger toxic effect of Cd. In conclusion, Enterobacter hormaechei and Priestia aryabhattai show high potential for use in bioremediation. One study, the growth rate and bioremediation potential of isolated strains resembling S. melonis and E. hormaechei were tested in a medium containing Ni, Cu, Pb, and Cd under four metal mixtures and at different pH values. It is reported that they achieved the highest heavy metal removal after 48 h at pH 6 (Heidari et al. 2020). Previous study results have also shown that Sphingomonas strains can degrade hazardous compounds (Heidari et al. 2020) such as pentachlorophenol (Yang and Lee 2008) and Cd (Tangaromsuk et al. 2002). It has been reported that E. hormaechei strains are potential bioremediators of contaminated soils and can be used as PGPR and phosphate solubilizing bacteria in agriculture (Fahsi et al. 2021). Moreover, in one study, the isolated bacterial strain Priestia aryabhattai, which possesses toxic metal healing properties, was isolated from contaminated soil and water, and described as a potential agent for arsenic remediation (Kumar et al. 2021). In addition, the isolate of Priestia megaterium, one of the bacteria isolated from sediments transported to the Black Sea with some toxic metal salts, is reported to provide important evidence of potential isolates for bioremediation and biotechnological approaches (Kalkan 2022).
In this study, a process using microorganisms was carried out to treat the potentially toxic metals in landfill leachate without pretreatment or posttreatment. However, over time, the leachate-sewage sludge will accumulate due to the activity of these microorganisms. Therefore, the elimination of heavy metal accumulation in leachate sludge is important to maintain the efficiency and effectiveness of the bio-sand filter system. For this reason, there is a need for studies that address the applicability of microbiological leaching (Tang et al. 2020), hyperaccumulating endophytes (Wang et al. 2021), or other chemical leaching (Zhang et al. 2020) methods. Recently, it has also been reported that the combination of physicochemical processes followed by biological pretreatments is very efficient, and it has been shown that both COD and N-NH4+ were almost completely removed by a combination of reverse osmosis and activated sludge (Tałałaj et al. 2019). More recently, bioleaching has been developed to recycle various metals in leachate or solid waste residues treated by filtration (Zeng et al. 2016; Dunbar 2017).
CONCLUSIONS
- Enterobacter hormaechei and mycobacteria are currently discussed in the literature for removing organic and inorganic pollutants. However, the efficiency of removal of toxic metals from wastewater, especially by Priestia aryabhattai strains, has yet to be discussed in the literature. Therefore, this study is unique and can be considered supportive of the literature concerning the other two isolated bacteria. Based on the research results, it is suggested that the effectiveness of these bacteria in removing heavy metals in the leachate at different pH, temperature, and contact time should be investigated in further field studies.
- The current study found that the isolated bacteria Enterobacter hormaechei and Priestia aryabhattai, adapted to environmental conditions, were very highly efficient in removing complicated, potentially toxic metals in the leachate at a rate of 68% and at 64%, respectively. Although, they have not been used for pre- or post-treatment of raw leachate. Therefore, it is supposed to be integrated or develop these bacterial sand filters with other conventional cost-effective systems to achieve safer wastewater treatment. Moreover, the researchers reported that this type of bacterial biofilter, besides its advantages, can be more than ten times cheaper than conventional wastewater treatment systems.
- Enterobacter hormaechei and Priestia aryabhattai have already been used to remove organic pollutants. However, the efficiency of removal of toxic metals in wastewater, especially by Priestia aryabhattai, has yet to be discussed in the literature. Therefore, this study can be considered original in this regard. For this reason, according to the research results, it is important to include these bacteria, whose efficiency in removing of heavy metals in leachate is studied, in new studies on the efficiency of removal of other pollutant parameters in wastewater issues.
ACKNOWLEDGEMENTS
The authors thank Karabuk University, which provides support with grant number KBUBAP-22-YL-031 and within the scope of for their support to the Scientific Research Unit.
STATEMENTS and DECLARATIONS
Availability of data and materials: Not applicable.
Funding: This research was funded by Karabuk University Coordinatorship of Research Projects, grant number KBUBAP-22-YL-031
Competing Interests: We know of no conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome. As Corresponding Author, I confirm that the manuscript has been read and approved for submission by all the named authors.
Ethics approval: Not applicable.
Consent to participate: Not applicable.
Consent for publication: Not applicable
Code availability: Not applicable.
Data availability: Not applicable
Author contributions: All authors: Conceptualization, Methodology, Data curation, Writing- Original draft preparation, Visualization. Investigation, Writing- Reviewing and Editing. Ayhan KOCAMAN: Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Visualization.
REFERENCES CITED
Abdolali, A., Guo, W., Ngo, H., Chen, S., Nguyen, N., and Tung, K. (2014). “Typical lignocellulosic wastes and by-products for biosorption process in water and wastewater treatment: A critical review,” Bioresource Technology 160, 57-66. DOI: 10.1016/j.biortech.2013.12.037
Adelopo, A., Haris, P. I., Alo, B., Huddersman, K., and Jenkins, R. (2018). “Multivariate analysis of the effects of age, particle size and landfill depth on heavy metals pollution content of closed and active landfill precursors,” Waste Management 78, 227-237. DOI: 10.1016/j.wasman.2018.05.040
Agency for Toxic Substances and Disease Registry (ATSDR) (2007). “CERCLA priority list of hazardous substances,” ATSDR, (http://www.atsdr.cdc.gov/cercla/07list.Html). Accessed 01 Jan 2022.
Ahalya, N., Ramachandra, T., and Kanamadi, R. (2003). “Biosorption of heavy metals,” Research Journal of Chemistry and Environment 7(4), 71-79.
Akgul, D., Aktan, C. K., Yapsakli, K., and Mertoglu, B. (2013). “Treatment of landfill leachate using UASB-MBR-SHARON–Anammox configuration,” Biodegradation 24, 399-412. DOI: 10.1007/s10532-012-9597-y
Al-Saadi, A. A., Saleh, T. A., and Gupta, V. K. (2013). “Spectroscopic and computational evaluation of cadmium adsorption using activated carbon produced from rubber tires,” Journal of Molecular Liquids 188, 136-142. DOI: 10.1016/j.molliq.2013.09.036
Amoako-Nimako, G.K., Chen, F., Fu, J., Yu, D., Yang, X. (2022). “The joint anaerobic denitrification performance of Klebsiella sp. and Enterobacter hormaechei using two carbon substrates with and without the presence of heavy metals,” Water, Air, & Soil Pollution 233(12), 531. DOI: 10.1007/s11270-022-05991-1
Awasthi, A., Pandey, A., and Khan, J. (2017). “Biosorption an innovative tool for bioremediation of metal-contaminated municipal solid waste leachate: Optimization and mechanisms exploration,” International Journal of Environmental Science and Technology 14, 729-742. DOI: 10.1007/s13762-016-1173-2
Azadi, D., Shojaei, H., Mobasherizadeh, S., Naser, A.D. (2017). “Screening, isolation and molecular identification of biodegrading mycobacteria from Iranian ecosystems and analysis of their biodegradation activity,” AMB Express 7, 1-15. DOI: 10.1186/s13568-017-0472-4
Chian, E. S., and DeWalle, F. B. (1976). “Sanitary landfill leachates and their treatment,” Journal of the Environmental Engineering Division 102(2), 411-431. DOI: 10.1061/JEEGAV.0000476
Davis-Hoover, W. J. (1990). Development of an in vivo Respiratory Model and its Use For Assessment of Infectivity of Heterotrophic Bacteria Isolated from Drinking Water, Ph.D. Dissertation, University of Cincinnati, USA.
Dhaka, V., Singh, S., Ramamurthy, P.C., Samuel, J., Swamy Sunil Kumar Naik, T., Khasnabis, S., Prasad, R., Singh, J. (2022). “Biological degradation of polyethylene terephthalate by rhizobacteria,” Environmental Science and Pollution Research 1-10. DOI: 10.1007/s11356-022-20324-9.
Dunbar, W. S. (2017). “Biotechnology and the mine of tomorrow,” Trends in Biotechnology 35(1), 79-89. DOI:10.1016/j.tibtech.2016.07.004
Dursun, A. Y. (2006). “A comparative study on determination of the equilibrium, kinetic and thermodynamic parameters of biosorption of copper (II) and lead (II) ions onto pretreated Aspergillus niger,” Biochemical Engineering Journal 28(2), 187-195. DOI: 10.1016/j.bej.2005.11.003
Esikova, T. Z., Anokhina, T. O., Abashina, T. N., Suzina, N. E., and Solyanikova, I. P. (2021). “Characterization of soil bacteria with potential to degrade benzoate and antagonistic to fungal and bacterial phytopathogens,” Microorganisms 9(4), 755. DOI: 10.3390/microorganisms9040755
Fahsi, N., Mahdi, I., Mesfioui, A., Biskri, L., and Allaoui, A. (2021). “Plant growth-promoting rhizobacteria isolated from the jujube (Ziziphus lotus) plant enhance wheat growth, Zn uptake, and heavy metal tolerance,” Agriculture 11(4), article 316. DOI: 10.3390/agriculture11040316
Falkinham 3rd, J., George, K., Parker, B., and Gruft, H. (1984). “In vitro susceptibility of human and environmental isolates of Mycobacterium avium, M. intracellulare, and M. scrofulaceum to heavy-metal salts and oxyanions,” Antimicrobial Agents and Chemotherapy 25(1), 137-139. DOI: 10.1128/aac.25.1.137
Fan, T., Liu, Y., Feng, B., Zeng, G., Yang, C., Zhou, M., Zhou, H., Tan, Z., and Wang, X. (2008). “Biosorption of cadmium (II), zinc (II) and lead (II) by Penicillium simplicissimum: Isotherms, kinetics and thermodynamics,” Journal of Hazardous Materials 160(2-3), 655-661. DOI: 10.1016/j.jhazmat.2008.03.038
Felsenstein, J. (1985). “Phylogenies and the comparative method,” The American Naturalist 125(1), 1-15. DOI: 10.1086/284325
Gotvajn, A. Ž., and Pavko, A. (2015). “Perspectives on biological treatment of sanitary landfill leachate,” Wastewater Treatment Engineering 13, 31-39. DOI: 10.5772/60924
Gupta, V. K. (1998). “Equilibrium uptake, sorption dynamics, process development, and column operations for the removal of copper and nickel from aqueous solution and wastewater using activated slag, a low-cost adsorbent,” Industrial & Engineering Chemistry Research 37(1), 192-202. DOI: 10.1021/ie9703898
Heidari, P., Sanaeizade, S., and Mazloomi, F. (2020). “Removal of nickel, copper, lead and cadmium by new strains of Sphingomonas melonis e8 and Enterobacter hormaechei WW28,” Journal of Applied Biotechnology Reports 7(4), 208-214. DOI: 10.30491/JABR.2020.120185
Hermosilla, D., Cortijo, M., and Huang, C. P. (2009). “Optimizing the treatment of landfill leachate by conventional Fenton and photo-Fenton processes,” Science of The Total Environment 407(11), 3473-3481. DOI: 10.1016/j.scitotenv.2009.02.009
Hussain, T. (2007). “Leprosy and tuberculosis: An insight-review,” Critical Reviews in Microbiology,” 33(1), 15-66. DOI: 10.1080/10408410601172271
Imron, M. F., Kurniawan, S. B., and Abdullah, S. R. S. (2021). “Resistance of bacteria isolated from leachate to heavy metals and the removal of Hg by Pseudomonas aeruginosa strain FZ-2 at different salinity levels in a batch biosorption system,” Sustainable Environment Research 31, article 14. DOI: 10.1186/s42834-021-00088-6
Imron, M. F., Kurniawan, S. B., and Soegianto, A. (2019). “Characterization of mercury-reducing potential bacteria isolated from Keputih non-active sanitary landfill leachate, Surabaya, Indonesia under different saline conditions,” Journal of Environmental Management 241, 113-122. DOI: 10.1016/j.jenvman.2019.04.017
Iravanian, A., and Ravari, S. O. (2020). “Types of contamination in landfills and effects on the environment: A review study,” IOP Conference Series: Earth and Environmental Science 614, article ID 012083. DOI: 10.1088/1755-1315/614/1/012083
Jayanthi, B., Emenike, C., Agamuthu, P., Simarani, K., Mohamad, S., and Fauziah, S. (2016). “Selected microbial diversity of contaminated landfill soil of Peninsular Malaysia and the behavior towards heavy metal exposure,” Catena 147, 25-31. DOI: 10.1016/j.catena.2016.06.033
Kalkan, S. (2022). “Heavy metal resistance of marine bacteria on the sediments of the Black Sea,” Marine Pollution Bulletin 179, article ID 113652. DOI: 10.1016/j.marpolbul.2022.113652
Kang, X., Zhao, X., Song, X., Wang, D., Shi, G., Duan, X., Chen, X., Shen, G. (2023). “Nitrogen removal by a novel strain Priestia aryabhattai KX-3 from East Antarctica under alkaline pH and low-temperature conditions,” Process Biochemistry. DOI:10.1016/j.procbio.2023.05.030
Kaur, S., and Roy, A. (2021). “Bioremediation of heavy metals from wastewater using nanomaterials,” Environment, Development and Sustainability 23, 9617-9640. DOI: 10.1007/s10668-020-01078-1
Kebabi, B., Aissaoui, S., Sifour, M., Ouled, H., Bougherara, H., and Aouati, M. (2018). “Heterotrophic denitrification by Enterobacter hormaechei collected from wastewater treatment plant in presence of methanol and sodium-succinate,” Journal of Materials and Environmental Sciences 9(3), 804-810. DOI: 10.26872/jmes.2018.9.3.88
Kim, S.-H., Kim, I. H., Lee, W. J., and Lee, J.-H. (2008). “Characterization of thiosulfate-oxidizing Enterobacter hormaechei JH isolated from barnyard manure,” Korean Journal of Chemical Engineering 25, 1131-1135. DOI: 10.1007/s11814-008-0185-0
King, H. C., Khera-Butler, T., James, P., Oakley, B. B., Erenso, G., Aseffa, A., Knight, R., Wellington, E. M., and Courtenay, O. (2017). “Environmental reservoirs of pathogenic mycobacteria across the Ethiopian biogeographical landscape,” PloS One 12(3), article ID e0173811. DOI: 10.1371/journal.pone.0173811
Kumar, P., Dash, B., Suyal, D. C., Gupta, S., Singh, A. K., Chowdhury, T., and Soni, R. (2021). “Characterization of arsenic-resistant Klebsiella pneumoniae RnASA11 from contaminated soil and water samples and its bioremediation potential,” Current Microbiology 78, 3258-3267. DOI: 10.1007/s00284-021-02602-w
Kumar, S., Stecher, G., and Tamura, K. (2016). “MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets,” Molecular Biology and Evolution 33(7), 1870-1874. DOI: 10.1093/molbev/msw054
Le, T. H., Hoang, Q. C., Vu, D. D., and Vo, T. H. T. (2021). “Biodegradation of organophosphorus insecticide methyl parathion by soil microorganisms,” E3S Web of Conferences 265, article 03002. DOI: 10.1051/e3sconf/202126503002
Le, T. H., Hoang, Q. C., Vu, D. D., and Vo, T. H. T. (2021). “Biodegradation of organophosphorus insecticide methyl parathion by soil microorganisms,” E3S Web of Conferences, EDP Sciences. pp. 03002. DOI: 10.1051/e3sconf/202126503002
Lu, H., Xia, C., Chinnathambi, A., Nasif, O., Narayanan, M., Shanmugam, S., Chi, N.T.L., Pugazhendhi, A., On-Uma, R., and Jutamas, K. (2023). “Evaluation of cadmium tolerance and remediated efficacy of wild and mutated Enterobacter species isolated from potassium nitrate (KNO₃) processing unit contaminated soil,” Chemosphere 311, article 136899. DOI: 10.1016/j.chemosphere.2022.136899
Masoner, J. R., Kolpin, D. W., Furlong, E. T., Cozzarelli, I. M., and Gray, J. L. (2016). “Landfill leachate as a mirror of today’s disposable society: Pharmaceuticals and other contaminants of emerging concern in final leachate from landfills in the conterminous United States,” Environmental Toxicology and Chemistry 35(4), 906-918. DOI: 10.1002/etc.3219
Mertens, D. (2005). “AOAC Official Method 922.02,” in: Plants Preparation of Laboratuary Sample. Official Methods of Analysis, 18th edn., W. Horwitz, and G. W. Latimer (Eds.) Gaitherburg, MD, USA, pp. 1-2.
Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics, Oxford University Press, Oxford, UK. DOI: 10.1017/S0016672301219405
Neneng, L., and Gunawan, Y. E. (2018). “The role of coenzymes on mercury (Hg2+) bioremediation by isolates Pseudomonas aeruginosa KHY2 and Klebsiella pneumonia KHY3,” Journal of Tropical Life Science 8(1), article ID 228158. DOI: 10.11594/jtls.08.01.04
Ozbay, G., Jones, M., Gadde, M., Isah, S., and Attarwala, T. (2021). “Design and operation of effective landfills with minimal effects on the environment and human health,” Journal of Environmental and Public Health 2021, article ID 6921607. DOI: 10.1155/2021/6921607
Park, A. J., Cha, D. K., and Holsen, T. M. (1998). “Enhancing solubilization of sparingly soluble organic compounds by biosurfactants produced by Nocardia erythropolis,” Water Environment Research 70(3), 351-355. DOI: 10.2175/106143098X124984
Rahman, Z., and Singh, V. P. (2019). “The relative impact of toxic heavy metals (THMs)(arsenic (As), cadmium (Cd), chromium (Cr)(VI), mercury (Hg), and lead (Pb)) on the total environment: An overview,” Environmental Monitoring and Assessment 191, article 419. DOI: 10.1007/s10661-019-7528-7
Razarinah, W., Zalina, M. N., and Abdullah, N. (2014). “Treatment of landfill leachate by immobilized Ganoderma australe and crude enzyme,” ScienceAsia 40, 335-339. DOI: 10.2306/scienceasia1513-1874.2014.40.335
Saetang, J., and Babel, S. (2012). “Biodegradation of organics in landfill leachate by immobilized white rot fungi, Trametes versicolor BCC 8725,” Environmental Technology 33(22), 2575-2584. DOI: 10.1080/09593330.2012.680917
Sanders, W. J., and Wolinsky, E. (1980). “In vitro susceptibility of Mycobacterium marinum to eight antimicrobial agents,” Antimicrobial Agents and Chemotherapy 18(4), 529-531. DOI: 10.1128/aac.18.4.529
Sağ, Y., and Kutsal, T. (1996). “The selective biosorption of chromium (VI) and copper (II) ions from binary metal mixtures by R. arrhizus,” Process Biochemistry 31(6), 561-572. DOI: 10.1016/S0032-9592(95)00100-X
Saitou, N., and Nei, M. (1987). “The neighbor-joining method: A new method for reconstructing phylogenetic trees,” Molecular Biology and Evolution 4(4), 406-425. DOI: 10.1093/oxfordjournals.molbev.a040454
Saleh, T. A. (2015a). “Isotherm, kinetic, and thermodynamic studies on Hg (II) adsorption from aqueous solution by silica-multiwall carbon nanotubes,” Environmental Science and Pollution Research 22, 16721-16731. DOI: 10.1007/s11356-015-4866-z
Saleh, T. A. (2015b). “Mercury sorption by silica/carbon nanotubes and silica/activated carbon: A comparison study,” Journal of Water Supply: Research and Technology—AQUA 64(8), 892-903. DOI: 10.2166/aqua.2015.050
Saleh, T. A., and Gupta, V. K. (2014). “Processing methods, characteristics and adsorption behavior of tire derived carbons: A review,” Advances in Colloid and Interface Science 211, 93-101. DOI: 10.1016/j.cis.2014.06.006
Satsuma, K., and Masuda, M. (2012). “Reductive dechlorination of methoxychlor by bacterial species of environmental origin: Evidence for primary biodegradation of methoxychlor in submerged environments,” Journal of Agricultural and Food Chemistry 60(8), 2018-2023. DOI: 10.1021/jf2048614
Tałałaj, I. A., Biedka, P., and Bartkowska, I. (2019). “Treatment of landfill leachates with biological pretreatments and reverse osmosis,” Environmental Chemistry Letters 17, 1177-1193. DOI: 10.1007/s10311-019-00860-6
Tangaromsuk, J., Pokethitiyook, P., Kruatrachue, M., and Upatham, E. (2002). “Cadmium biosorption by Sphingomonas paucimobilis biomass,” Bioresource Technology 85(1), 103-105. DOI: 10.1016/S0960-8524(02)00066-4
Tang, J., He, J., Tang, H., Wang, H., Sima, W., Liang, C., and Qiu, Z. (2020). “Heavy metal removal effectiveness, flow direction and speciation variations in the sludge during the biosurfactant-enhanced electrokinetic remediation,” Separation and Purification Technology 246, article 116918. DOI: 10.1016/j.seppur.2020.116918
Tunali, S., and Akar, T. (2006). “Zn (II) biosorption properties of Botrytis cinerea biomass,” Journal of Hazardous Materials 131(1-3), 137-145. DOI: 10.1016/j.jhazmat.2005.09.024
Valdiviezo, A., Luo, Y.-S., Chen, Z., Chiu, W. A., and Rusyn, I. (2021). “Quantitative in vitro-to-in vivo extrapolation for mixtures: A case study of superfund priority list pesticides,” Toxicological Sciences 183(1), 60-69. DOI: 10.1093/toxsci/kfab076
Vijayaraghavan, K., and Balasubramanian, R. (2015). “Is biosorption suitable for decontamination of metal-bearing wastewaters: A critical review on the state-of-the-art of biosorption processes and future directions,” Journal of Environmental Management 160, 283-296. DOI: 10.1016/j.jenvman.2015.06.030
Vijayaraghavan, K., and Yun, Y.-S. (2008). “Bacterial biosorbents and biosorption,” Biotechnology Advances 26(3), 266-291. DOI: 10.1016/j.biotechadv.2008.02.002
Vodyanitskii, Y. N. (2016). “Biochemical processes in soil and groundwater contaminated by leachates from municipal landfills: (Mini review),” Annals of Agrarian Science 14(3), 249-256. DOI: 10.1016/j.aasci.2016.07.009
Volesky, B. (2007). “Biosorption and me,” Water Research 41(18), 4017-4029. DOI: 10.1016/j.watres.2007.05.062
von Sperling, M. (1996). “Comparison among the most frequently used systems for wastewater treatment in developing countries,” Water Science and Technology 33(3), 59-72. DOI: 10.1016/0273-1223(96)00301-0
Wang, J., and Chen, C. (2009). “Biosorbents for heavy metals removal and their future,” Biotechnology Advances 27(2), 195-226. DOI: 10.1016/j.biotechadv.2008.11.002
Wang, S., Liu, T., Xiao, X., and Luo, S. (2021). “Advances in microbial remediation for heavy metal treatment: a mini review,” Journal of Leather Science and Engineering 3, 1-10. DOI: 10.1186/s42825-020-00042-z
Yang, C.-F., and Lee, C.-M. (2008). “Pentachlorophenol contaminated groundwater bioremediation using immobilized Sphingomonas cells inoculation in the bioreactor system,” Journal of Hazardous Materials 152(1), 159-165. DOI: 10.1016/j.jhazmat.2007.06.102
Yetunde Mutiat, F.-B., Gbolahan, B., and Olu, O. (2018). “A comparative study of the wild and mutated heavy metal resistant Klebsiella variicola generated for cadmium bioremediation,” Bioremediation Journal 22(1-2), 28-42. DOI: 10.1080/10889868.2018.1445695
Zeng, J., Gou, M., Tang, Y.-Q., Li, G.-Y., Sun, Z.-Y., and Kida, K. (2016). “Effective bioleaching of chromium in tannery sludge with an enriched sulfur-oxidizing bacterial community,” Bioresource Technology 218, 859-866. DOI:10.1016/j.biortech.2016.07.051
Zhang, H., Yuan, X., Xiong, T., Wang, H., and Jiang, L. (2020). “Bioremediation of co-contaminated soil with heavy metals and pesticides: Influence factors, mechanisms and evaluation methods,” Chemical Engineering Journal 398, article ID 125657. DOI: 10.1016/j.cej.2020.125657
Zhang, L., Chen, Y., Ma, C., Liu, L., Pan, J., Li, B., Wu, X., amd Wang, Q. (2020). “Improving heavy metals removal, dewaterability and pathogen removal of waste activated sludge using enhanced chemical leaching,” Journal of Cleaner Production 271, article 122512. DOI: 10.1016/j.jclepro.2020.122512
Article submitted: April 25, 2023; Peer review completed: June 17, 2023; Revised version received and accepted: June 26, 2023; Published: June 30, 2023.
DOI: 10.15376/biores.18.3.5476-5493
