Abstract
Manganese peroxidase (MnP), a crucial enzyme in biodegradation of lignin, is synthesized by most white rot fungi. To obtain novel enzymes with superior biodegradation potential, MnP-producing wild isolates were evaluated for their ability to degrade recalcitrant azo dyes, sulfonephthalein dyes, and kraft lignin. Of 30 wild isolates screened, 18 tested positive for lignin modifying enzymes (LMEs). Thirteen of these isolates were positive for both laccase and MnP, whereas four produced only laccase, and one produced lignin peroxidase alone. The isolates were identified as Clitopilus scyphoides MH172162 (AGUM004), Ganoderma rasinaceum MH172163 (AGUM007), and three Schizophyllum species: MH172164, MH172165, and MH172166 (KONA001, AGUM0011, and AGUM021). The Fourier-transform infrared spectroscopy (FTIR) analysis of dye degradation and kraft lignin samples with AGUM004 and AGUM007 revealed biotransformation. The former could not completely degrade Reactive Black 5 and bromocresol green, but it could completely (100%) decolorize bromophenyl blue, bromothymol blue, and Remazol brilliant blue R. The latter efficiently degraded almost all tested dyes. Both degraded kraft lignin. The screened hyper MnP-producing wild AGUM004 and AGUM007 were shown to be potential dye degraders in addition to having lignin degrading abilities.
Download PDF
Full Article
Screening of Wild Basidiomycetes and Evaluation of the Biodegradation Potential of Dyes and Lignin by Manganese Peroxidases
Ramya G. Rao,a,b Aarthi Ravichandran,a Giridhar Kandalam,c Samanta Ashish Kumar,d Senani Swaraj,d and Manpal Sridhar a,*
Manganese peroxidase (MnP), a crucial enzyme in biodegradation of lignin, is synthesized by most white rot fungi. To obtain novel enzymes with superior biodegradation potential, MnP-producing wild isolates were evaluated for their ability to degrade recalcitrant azo dyes, sulfonephthalein dyes, and kraft lignin. Of 30 wild isolates screened, 18 tested positive for lignin modifying enzymes (LMEs). Thirteen of these isolates were positive for both laccase and MnP, whereas four produced only laccase, and one produced lignin peroxidase alone. The isolates were identified as Clitopilus scyphoides MH172162 (AGUM004), Ganoderma rasinaceum MH172163 (AGUM007), and three Schizophyllum species: MH172164, MH172165, and MH172166 (KONA001, AGUM0011, and AGUM021). The Fourier-transform infrared spectroscopy (FTIR) analysis of dye degradation and kraft lignin samples with AGUM004 and AGUM007 revealed biotransformation. The former could not completely degrade Reactive Black 5 and bromocresol green, but it could completely (100%) decolorize bromophenyl blue, bromothymol blue, and Remazol brilliant blue R. The latter efficiently degraded almost all tested dyes. Both degraded kraft lignin. The screened hyper MnP-producing wild AGUM004 and AGUM007 were shown to be potential dye degraders in addition to having lignin degrading abilities.
Keywords: Manganese peroxidase; Clitopilus scyphoides; Ganoderma rasinaceum; Biodegradation; Dye decolorization; Lignin
Contact information: a: Bioenergetics and Environmental Sciences Division, ICAR-National Institute of Animal Nutrition and Physiology, Adugodi, Bengaluru 560 030, India; b: PhD Scholar, Department of Biochemistry, Jain University, Bengaluru, 560011, Karnataka, India; c: Knowledge management and Biostatistics Section, ICAR-National Institute of Animal Nutrition and Physiology, Adugodi, Bangalore, 560 030, Karnataka, India; d: Animal Nutrition Division, ICAR-National Institute of Animal Nutrition and Physiology, Adugodi, Bangalore, 560 030, Karnataka, India;
* Corresponding author: manpalsridhar@ yahoo.co.uk
INTRODUCTION
Fungi play important roles in biodegradation and bioremediation. White rot fungi (WRF), a group within the class Basidiomycetes, have the unique ability to degrade and mineralize recalcitrant hetero-polymer lignin, which is the most abundant organic material on Earth (Erden et al. 2009; Cesarino et al. 2012). White rot fungi are ubiquitous in nature and grow as saprophytes on dead and decaying trees, usually in the forest ecosystem, and are exceptional in their ability to degrade lignin selectively. They possess a set of nonspecific enzymes called lignin modifying enzymes (LME), which degrade lignin and other recalcitrant compounds such as munitions waste, pesticides, polychlorinated biphenyls, polycyclic aromatic hydrocarbons, bleach plant effluent, synthetic dyes, synthetic polymers, and wood preservatives (Pointing 2001). These LME are a set of extracellular enzymes secreted by WRF, and are mainly comprised of laccase, manganese peroxidase (MnP), lignin peroxidase (LiP), and versatile peroxidase (VP), along with oxidases, which generate extracellular H2O2. Different species of white rot fungi may secrete one, two, or all of these enzymes (Wesenberg et al. 2003).
Within the LME, MnP is synthesized by almost all the lignin-degrading WRF and is the most crucial in lignin biodegradation. The MnPs are peroxide-dependent extracellular glycoproteins with one molecule of heme (Dashtban et al. 2010). In the presence of hydrogen peroxide, they oxidize Mn (II) to highly reactive Mn (III). Mn (III) further chelates with organic acids, which attack and oxidize lignin and other recalcitrant compounds (Van Aken and Agathos 2002). Since the discovery of MnP in P. chrysosporium (Glenn and Gold 1985), MnPs have been found in other basidiomycetes, such as Bjerkandera sp. strain BOS55 (Mester and Field 1997), Bjerkandera sp. (Palma et al. 2000), Agaricus bisporus (Lankinen et al.2001), Lenzites betulinus (Hoshino et al. 2002), Phanerochaete flavido-alba (de la Rubia et al. 2002), Phanerochaete sordida (Harazonoet al. 2003), Panus tigrinus (Lisov et al. 2003), Ganoderma lucidium (Novotnýet al. 2004), Dichomitus squalens (Eichlerová et al. 2005), Lentinula edodes (Boer et al. 2004), Nematoloma frowardii b19 (Hildén et al. 2008), Pleurotus pulmonarius (dos Santos Bazanella et al. 2013), Trametes versicolor, Pleurotus ostreatus, Irpex lacteus (Zhao et al. 2015), and Trametes hirsuta (Vasina et al. 2017).
The potential applications of MnPs rival those of laccases. They degrade both phenolic and non-phenolic lignin model compounds (Wang et al. 2018). They are efficient degraders of many recalcitrant compounds including different synthetic dyes (Qin et al. 2014), herbicides, polycyclic aromatic hydrocarbons, mycotoxins, estrogens, explosives, and antifouling compounds (Wang et al. 2018). The presence of MnP increases the degree of dye decolorization (Chagas and Durrant 2001; Christian et al. 2003).
Considering the wide application of MnPs, the current study was conducted to screen for wild WRF exhibiting higher MnP activities and identify their phylogenetic relationships. The wild isolates producing greater quantities of MnP were further evaluated for their ability to degrade recalcitrant azo dyes and sulfonephthalein dyes. They were tested for the biodegradation of kraft lignin. The extent of biodegradation of different dyes and kraft lignin was measured using spectrophotometry, and patterns of degradation were studied using Fourier-transform infrared spectroscopy (FTIR) techniques. The identified isolates producing MnP have not been explored previously.
EXPERIMENTAL
Material and Methods
Chemicals
Chemicals used in the study were of analytical grade unless otherwise stated. Reactive Black 5 (RB5), 2,2’-azino-bis(3-ethyl benzothiazoline-6-sulfonic acid) (ABTS), and Remazol brilliant blue R (RBBR) used for assay were procured from Sigma Aldrich (St. Louis, MO, USA).
Collection, isolation, and maintenance of white rot fungi
Fruiting bodies of 30 different WRF were collected in clean dry self-sealing polythene bags from forests in Agumbe, Madikere and in and around Shimoga (Karnataka State), India. In all cases, the substrate was represented by wood found in various stages of decay.
The cultures were marked with information such as number and isolation location. Pure cultures from collected samples were obtained by tissue culture technique (Pradeep et al. 2013). All pure cultures were maintained on PDA slants and stored at 4 °C for further use.
Screening for LME production
Initial screening for lignolytic enzyme production was carried out on 0.02% guaiacol-supplemented PDA. All cultures were inoculated on pre-solidified PDA medium supplemented with guaiacol and incubated for 7 days at room temperature to check the phenolic oxidation. The positive cultures from primary screening were then grown in basal medium which contained 0.1% KH2PO4, 0.05% C4H12N2O6, 0.05% MgSO4.7H2O, 0.001% of CaCl2 and yeast extract, and 0.0001% of CuSO4.5H2O, Fe2(SO4)3, and MnSO4.H2O. Fungal cultures pre-grown in PDB were used as inoculum. The cells were homogenized prior to inoculation, and 2% of culture suspension was added to the production medium. Flasks were maintained under continuous shaking at 80 rpm at 30 C for 7 days. Culture filtrates obtained were assessed for the production of laccase and other peroxidases. All experiments were performed in triplicate.
Enzyme Assay
Culture filtrate was used as the enzyme source to determine the ligninolytic enzymes activity. A sample of 500 µL was taken to check different enzyme production. Laccase production was assessed by a measurement of the enzymatic oxidation of ABTS at 420 nm (Kinnunen et al. 2016).
The reaction mixture contained 0.8 mM ABTS, 0.4 M sodium acetate buffer (pH5.2), and 500 µL of 0.5 mg/mL catalase. The MnP activity was assayed by the oxidation of 4 mM MnSO4in 50 mM sodium malonate buffer (pH 4.5), in the presence of 0.4 mM H2O2. Oxidation of MnSO4 was measured by increase in OD at 270 nm (Wariishi et al. 1992).
Lignin peroxidase was determined by Azure B assay (Archibald 1992). Reaction mixture for assay contained 0.04 mM Azure B, 100 mM sodium tartarate (pH 4.5), and 0.4 mM H2O2. Oxidation of Azure B was determined by the decrease in OD at 651 nm. Production of versatile peroxidase was assessed by RB5 assay (Jarosz-Wilkołazka et al. 2009). Oxidation of RB5 was determined in 100 mM sodium tartrate buffer at pH 3 with 10 µM of RB5. The reaction was initiated by addition of 0.1mM H2O2 and assessed through decrease in absorbance at 598 nm.
Identification and Phylogenetic Analysis of WRF
The proficient strains unveiling hyper MnP activity were identified in a separate study carried out by Rao et al. (2018). Genomic DNA of selected WRF were isolated by the microwave method, which was then amplified using PCR with ITS 1 (5′-TCCGTAGGTGAACCTGCG G- 3′) and ITS 4 (5′-TCCTCCGCTTATTGATAT G-3′) as forward and reverse primers, respectively. The amplified product comprised of 18s rDNA, 28s rDNA, and5.8s rDNA, along with ITS1 and ITS4 regions, was sequenced. The ITS sequence obtained was then subjected to sequence comparison through BLAST. Culture names were assigned based on more than 99% sequence similarity.
To study the morphology of identified isolates, culture stained with lacto-phenol blue dye, was examined under microscope. The morphology of the fungus and details of hyphae structure and spores were observed at 60x magnification (Pradeep et al. 2013).
The identified isolates were studied for their evolutionary relationship with their phylogeny using MEGAX software (Kumar et al. 2018). A total of 44 related species’ DNA sequences comprised of 18s rDNA, 28s rDNA, and5.8s rDNA, along with ITS1 and ITS4 regions, were selected; the data were retrieved from NCBI. Maximum likelihood statistical method was selected to compute the evolutionary distance (Tamura et al. 2004). Bootstrap method with 1000 replicates was adopted to find the phylogenetic evaluation (Felsenstein1985). To construct the phylogenetic tree, Nearest-Neighbor-Interchange (NNI) was considered (Saitou and Nei 1987). All positions containing gaps and missing data were eliminated.
MnP Production
The biosynthesis trend of MnP from AGUM004 and AGUM007 was studied using modified production media (Takano et al. 2004). Production media was composed of 0.4g K2HPO4, 0.6 g KH2PO4, 0.5 g MgSO4, 0.03 g MnSO4, 0.01 g CaCO3, 1 g NH4H2PO4, 0.25 g yeast extract, 10 g glucose, 0.5 g Tween 80, and 8.3 g sodium malonate in 1000 mL of distilled water. Then 50 mL of production media in 250-mL shaken flasks were inoculated with 2% well grown culture in LME basal medium and incubated for 7 days at 30 ºC with constant shaking at 120 rpm. Activities of MnP and laccase were monitored from 0 h to 10 days at intervals of 24 h. Experiment was carried out in 5 replicates.
Dye Decolorization and Lignin Degradation Studies
To study the dye decolorization and delignification proficiency, hyper MnP-producing strains AGUM004 and AGUM007 were considered. For this, 20 mL of MnP production medium supplemented with 0.02% of RBBR, RB5, bromocresol green, bromothymol blue, and kraft lignin in 100 mL shaken flasks were inoculated with 2% well grown culture in LME basal medium and incubated for 7 days at 30 °C with constant shaking at 120 rpm. Uninoculated flasks with dyes/lignin were considered as control. Culture filtrates were examined on a spectrophotometer at the absorbance maxima for different dyes and lignin. Degradation efficacy of the culture was determined in terms of percentage of color removal and was calculated according to the formula given below,
(1)
where C is the current concentration of dye in a control sample (mg/L) and S is the current residual concentration of dye in the samples. Results were expressed in % degradation.
All the samples were also analyzed for dye and lignin breakdown pattern by using the FTIR technique. The FTIR analysis was carried out in the mid IR region of 400 cm-1 to 4000 cm-1. Graphs were reformed using Originpro software for comparison and interpretation. Absorption spectra in the region between 3100 cm-1 and 3600 cm-1 indicated the presence of exchangeable protons, typically from alcohol, amine, amide, or carboxylic acid groups, which was of less importance in interpreting the data (Coates 2000). Therefore, the region above 3000 cm-1 was omitted in the reformed graphs. The FTIR analysis was outsourced from SITC, University of Science and Technology, Cochin, India.
RESULTS AND DISCUSSION
Screening for LME Production
The 30 WRF were collected together to check the production of ligninolytic enzymes. Out of 30 cultures, 18 cultures were found to be positive for guaiacol oxidation in the primary screening, which showed an intense brown halo in and around the culture and indicated the production of ligninolytic enzymes, while 40% of the culture specimens failed to develop brown color around the mycelial growth (Fig. 1). Further investigation on lignolytic enzymes production in liquid medium, revealed that 13 WRF were both MnP and laccase producers, 4 cultures were only laccase producers, and only 1 culture was a lignin peroxidase producer. However, VP was not detected in any of the culture filtrates. Out of 13 MnP positive cultures, AGUM004 and AGUM007 were observed to secrete higher quantities of both MnP and laccase enzymes. Table 1 specifies the morphological appearance of positive cultures on PDA along with ability of guaiacol oxidation and lignolytic enzyme quantification.
Fig. 1. Guaiacol oxidation: control (uninoculated plate) (A); positive for guaiacol oxidation (B); positive culture on PDA without guaiacol (C); negative for guaiacol oxidation (D)
Identification and Phylogenetic Analysis of WRF
As AGUM004, AGUM007, KONA001, AGUM011, and AGUM021 were showing greater MnP activity, they were considered further for culture identification as well as phylogenetic studies. Isolates were identified as Clitopilus scyphoides (AGUM004), Ganoderma rasinaceum (AGUM007), and theother three as Schizophyllum species (KONA001, AGUM0011, and AGUM021).
Table 1. Morphological Appearance of WRF along with Primary Screening Result (Guaiacol reaction) and Quantification of their Lignolytic Enzymes
Fig. 2. Microscopic observation of (A) AGUM004; (B) AGUM007; (C) KONA001; (D) AGUM011; and (E) AGUM021
The PCR amplified DNA sequence through ITS1 and ITS4 primers were of 700 bp length comprised of ITS1, ITS2, and rDNA regions, which were deposited to Genbank with accession numbers MH172162 (Clitopilus scyphoides isolate AGUM004), MH172163 (Ganoderma rasinaceum isolate AGUM007), MH172164 (Schizophyllum sp. isolate KONA001), MH172165 (Schizophyllum sp. isolate AGUM011), and MH172166 (Schizophyllum commune isolate AGUM021). Figure 2 reveals microscopic observation of the type of hyphae, septa, clamp connections, and the features of the spores, which further confirmed these isolates belong to basidiomycetes. Morphological details revealed that Clitopilus scyphoides had septate hyphae and elliptical spores (Fig. 2A), Ganoderma rasinaceum formed typical almond-shaped basidiospores unique to the genus with septate hyphae (Fig. 2B), and the other three isolates of Schizophyllum had simple septate hyphae forming clamps with absence of basidiospores (Figs. 2C, 2D, and 2E).
Fig. 3. Phylogenetic tree of identified wild WRF revealing the phylogenic evolution
Phylogenetic analysis of all 5 isolates, along with their related species, was carried out to obtain evolutionary relationships of taxa. The optimal tree with the sum of branch length = 2.29447280 was shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Fig. 3).
The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. This analysis involved 43 nucleotide sequences. There were 932 positions in the final dataset. Phylogenetic analysis confirmed the isolates belonged to genera Clitopilus (AGUM004), Ganoderma (AGUMU007), and Schizophyllum (KONA001, AGUM011, and AGUM021).
MnP Production
Production trial of MnP from AGUM004 and AGUM007 revealed that production of MnP biosynthesis started from day 3, reached maximum on day 5, and started to decline thereafter. Laccase biosynthesis was observed from day 1, reached maximum on day 3, and then declined drastically thereafter. Maximum MnP produced was 1890±109 U/L from AGUM004 and 1623±152 U/L from AGUM007. Figure 4 shows the activity trend of both MnP and laccase from day 0 to day 7 in intervals of 24 h.
Fig. 4. Production of MnP (______) and laccase (_ _ _) by C. scyphoides (A) and G. rasinaceum (B) using MnP production media
Dye Decolorization and Lignin Degradation Studies
AGUM004 and AGUM007 were further considered for the biodegradation study. Both isolates exhibited different levels of lignin biodegradation and decolorization of different dyes.
Though AGUM004 could not completely degrade bromocresol green and RB5, it could completely (100%) decolorize dyes such as bromophenyl blue, bromothymol blue, and RBBR, whereas AGUM007 could efficiently (100%) degrade almost all the dyes including bromophenol blue, bromothymol blue, bromocresol green, and RBBR, but could only partially degrade RB5 (Table 2). Both AGUM004 and AGUM007 could degrade kraft lignin up to 46% and 48% only.
The FTIR analysis of dye degradation and kraft lignin samples revealed the biotransformation of the dyes and lignin, which reflected the spectrophotometric results. The FTIR of control and biodegraded samples was compared for major and fingerprint spectra (Table 3).
Table 2. Percent Degradation of Different Compounds by AGUM004 and AGUM007
Table 3. List of Changes Obtained in the Major Finger Print Spectra of Different Compounds by AGUM004 and AGUM007 Compared with Control
† Values are presented in mean ± standard deviation where ever applicable
Considerable changes in the peaks of spectra were observed for the biodegraded samples (Fig. 5Ato Fig. 5G). In the graph 5A-C for all the sulfonephthalein dyes, both alknyl C=C stretching at around 2300 cm-1, aromatic C=C bending at around 1600 cm-1, C=O bending at around 1400 cm-1, vibration of sulfonate groups between 1000 cm-1 and 1200 cm-1, and stretching of C-Br at 580 cm-1 were observed in the control samples. For both AGUM004 and AGUM007, peaks at around 2300 cm-1 and at around 1400 cm-1 were absent, a few spectra between 1000 cm-1 and 1200 cm-1 had disappeared, and shifts in the peaks for C-Br at 580 cm-1 were noticed. The comparison of FTIR spectra of control RB5 dye with degraded dye by AGUM007 and AGUM004 (Fig. 5D) revealed that AGUM004 failed to degrade the dye, whereas AGUM007 had degraded the azo bond and sulfonate groups, which was clearly indicated by the removal of bands near 1400 cm-1 and 1200 cm-1, respectively. For RBBR, an anthraquinone dye, the characteristic bands (Fig. 5E) were observed around 1600 cm-1 wavenumber for C=C bending in benzene ring and between 1100 cm-1 and 1200 cm-1 for vibration of sulfonate groups in the control samples. In samples of AGUM007 and AGUM004, the sulfonate groups were removed, which could be confirmed by the bands for sulfonate groups between 1100 cm-1 to 1200 cm-1 that disappeared. Further drastic change in absorption pattern for aromatic C=C was observed for both RBBR samples of AGUM007 and AGUM004. For RB5, an azo dye, spectra were assigned at around 2400 cm-1 for alknyl C=C stretching, at around 1600 cm-1 for C=C stretching, between 1100 cm-1 and 1200 cm-1 for vibration of sulfonate groups, and 1400 cm-1 for characteristic azo bands (N=N stretch) (Kalkan et al. 2014). For kraft lignin, peaks were assigned at 1771 cm-1 for aromatic C=O, 1700 cm-1 to 1750 cm-1 for unconjugated ketones, carbonyls, and ester groups; 1722 cm-1 for aliphatic C=O, from 1650 cm-1to 1680 cm-1 for conjugated p-substituent carbonyl and carboxyl C=O, 1500 cm-1 to 1600 cm-1, and 1420 cm-1 to 1430 cm-1 for benzene ring aromatic skeletal, 1325cm-1 to 1330 cm-1, and 1230 cm-1 to 1235 cm-1 for C-O of syringyl ring, 1270 cm-1 to 1275 cm-1 for C-O of guaiacyl ring, 1140 cm-1 to 1145cm-1 for C-H of guaiacyl ring, 1130 cm-1 for CH of syringyl ring, 1085 cm-1 to 1090 cm-1 for C-O of secondary alcohol and aliphatic ether, 1025 cm-1 to 1145 cm-1 for C-H of aromatic and primary alcohol, and 750 cm-1 to 860 cm-1 for C-H of aromatic ring (Lu et al. 2017).
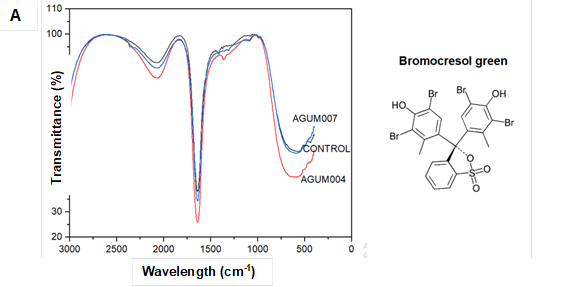
Fig. 5. FTIR spectra for different dyes and kraft lignin
Comparison of FTIR spectra of kraft lignin (Fig.5F) revealed that AGUM007 could transform peaks around 1230 cm-1 to 1235 cm-1; 1130 cm-1; 1085 cm-1 to 1090 cm-1; 750 cm-1 to 860 cm-1; as well as at 1771 cm-1 ,compared to the control whereas, in the case of AGUM004 major changes were observed only in peaks around 750 cm-1 to 860 cm-1 and 1771 cm-1.
MnP acts as an effective biochemical tool in degrading a variety of recalcitrant compounds with the help of reactive Mn(III) (Hofrichter 2002). The study results showed that 72% of guaiacol oxidation-positive isolates were MnP producers. Guaiacol is the most widely-used indicator compound for screening LME-producing WRF when compared to other compounds including tannic acid, gallic acid, syringaldehyde, etc., which directly indicates the presence of LME (Kim et al. 2010; Bodke et al. 2012; Pollegioniet al. 2015). Primary qualitative assay always helps in screening of large number of isolates as it consumes less time and is easy to interpret.
Naturally, WRF produces more than one LME or an array of LMEs to degrade lignin in the wood, and oxidation of guaiacol alone does not indicate the presence of any one particular enzyme. Therefore, quantitative estimation of LME was carried out using LME basal medium. Quantitative analysis of culture filtrate by isolated culture revealed that among positive isolates, majorities were either laccase producers or both laccase and MnP producers. Among the MnP-positive cultures, AGUM004 and AGUM007 were found to be hyper MnP-producing isolates with 111 U/L and 99 U/L activities, respectively. The quantitative analysis of the ligninolytic enzymes secreted by these wild isolates confirmed the presence of MnP, and further microscopic observation of spores, hyphae, and septa confirmed the isolates belonged to the basidiomycetes group. Yet molecular identification techniques involving DNA sequences are more precise and efficient. Consequently, the more frequently employed method of using Internal Transcribed Sequence (ITS) region of ribosomal DNA through the use of ITS 1 and ITS 4 primers to identify unknown fungal isolates was adopted. The isolates were identified through comparison with BLAST of the known fungal sequences in a distinct study carried out in the authors’ laboratory. However, the phylogenetic analysis did not confirm the novel isolates to their species level, yet confirmed their respective genera.
Strains of Clitopilus scyphoides and Ganoderma rasinaceum are scarcely reported in the literature as MnP and laccase producers. The presence of MnP along with laccase has been reported in Ganoderma lucidium strains (D’souza et al. 1999; Songulashvili et al. 2008),Pleurotus strains (Patrick et al. 2011; Camassola et al. 2013), Echinodontium taxodii 2538 (Kong et al. 2016), Trametes versicolor (Champagne and Ramsay 2005), and Panus tigrinus (Maltseva et al. 1991).
Production of MnP under submerged conditions drastically increased the production of MnP in the culture media. Production of laccase in the same media, though evident from day 1 itself, reached maximum on day 3, and started to decline subsequently when production of MnP started exponentially. MnSO4, Tween 80, and sodium malonate are inducers for MnP biosynthesis in different WRF (Perez and Jeffries 1992; Acevedo et al. 2011; Järvinen et al. 2012; Usha et al. 2014). Production of laccase did not drastically increase unlike MnP in the media used. This could be explained by the requisite of copper ions supplementation in the media for the production of higher levels of laccase (Baldrian 2003; Ire and Ahuekwe 2016).
White rot fungi are efficient degraders of recalcitrant synthetic dyes because of their nonspecific enzyme system (Murugesan and Kalaichelvan 2003). The ability to degrade different dyes has been observed in Pleurotus ostreatus (Palmieri et al. 2005; Shrivastava et al. 2005; Erkurt et al. 2007), Phanerochaete chrysosporium (Christian et al. 2003), Coriolus versicolor and Funalia trogii (Erkurt et al. 2007), and Cerrena unicolor BBP6 (Zhang et al. 2018). In most of the studies, WRF took more than 10 days to degrade the dyes, but isolates AGUM007 and AGUM004 in the study were more efficient and could degrade almost all the compounds within 7 days from the day of incubation. These findings are in agreement with Patrick et al. (2011), who found laccase and MnP together in Pleurotus sajorcaju could degrade RBBR within 7 days. Though MnP is known to degrade sulfonephthalein dyes by using free phenolic moiety present in SP dyes, which oxidizes to form carbon-centered cation by enzyme-generated Mn3+ (Christian et al. 2003), it is always a multi-enzyme action that is more efficient in biodegradation and bioremediation, which reflects the physiological conditions of basidiomycetes in action (Patrick et al. 2011). To see the pattern of degradation of dyes by the isolates screened, all the samples were analyzed for FTIR. The FTIR method acts as a valuable tool in dye decolorization studies, as it helps determine the type and strength of interactions occurring in the different functional groups of dyes after bio-transformation by microorganisms. Even a small shift in the spectra, deletion and/or addition of peaks determines the biotransformation of a compound (Coates 2000). The FTIR spectra of different dyes incubated with AGUM004 and AGUM007 in the current study have shown variations when compared with control samples, which in turn confirmed the degradation of dyes.
A number of studies have supplemented enough proof that lignin degradation in different straws with fungal pretreatment is more effective in improving the feed quality as compared to bacterial pretreatment (Madadi and Abbas 2017). To test the proficiency of isolates AGUM004 and AGUM007 in lignin degradation, degradation of kraft lignin was studied. The FTIR results showed that both isolates were equally capable of degrading kraft lignin. When the spectra of kraft lignin samples were compared, it was revealed that AGUM007 had degraded most of C-O in syringyl ring, C-O in guaiacyl ring, C-H in guaiacyl ring, and CH in syringyl ring, along with aromatic C=O and aromatic C-H bonds, whereas AGUM004 could majorly degrade aromatic C=O and aromatic C-H bonds. Similar findings were reported by Sahadevan et al. (2016) and Yang et al. (2010).
CONCLUSIONS
- Screened wild isolates exhibiting higher MnP activities were identified as Clitopilus scyphoides (AGUM004), Ganoderma rasinaceum (AGUM007), and Schizophyllum species (KONA001, AGUM0011, and AGUM021).
- Maximum MnP activity of 1890±109 U/L and 1623±152 U/L was produced by Clitopilus scyphoides and Ganoderma rasinaceum and was found to be highly efficient in degrading of various sulfonephthalein, azo, and anthraquinone dyes as well as kraft lignin.
- Because AGUM004 (Clitopilus scyphoides), and AGUM007 (Ganoderma rasinaceum) are potential dye degraders in addition to having lignin degrading ability, they promise wide range of industrial, biotechnological, and environmental applications.
ACKNOWLEDGMENTS
The financial assistance from the Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No.BT/PR11205/AAQ/1/589/2014) to perform the present research is gratefully acknowledged. The authors thank the Director of ICAR National Institute of Animal Nutrition and Physiology, Bangalore (India) for providing facilities to perform the research work.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest regarding the material discussed in the manuscript.
REFERENCES CITED
Acevedo, F., Pizzul, L., Castillo, M. P., Rubilar, O., Lienqueo, M. E., Tortella, G., and Diez, M. (2011). “A practical culture technique for enhanced production of manganese peroxidase by Anthracophyllum discolor Sp4,” Braz. Arch. Biol. Technol. 54(6), 1175-1186. DOI: 10.1590/S1516-89132011000600013
Archibald, F. S. (1992). “A new assay for lignin-type peroxidases employing the dye azure B,” Appl. Environ. Microbiol. 58(9), 3110-3116.
Baldrian, P. (2003). “Interactions of heavy metals with white-rot fungi,” Enzyme Microb. Technol. 32(1),78-91. DOI: 10.1016/S0141-0229(02)00245-4
Bodke, P. M., Senthilarasu, G., and Raghukumar, S. (2012). “Screening diverse fungi for laccases of varying properties,” Indian J. Microbiol. 52(2), 247-250. DOI: 10.1007/s12088-011-0204-4
Boer, C. G., Obici, L., de Souza, C. G. M., and Peralta, R. M. (2004). “Decolorization of synthetic dyes by solid state cultures of Lentinula (Lentinus) edodes producing manganese peroxidase as the main ligninolytic enzyme,” Bioresour. Technol. 94(2), 107-112.DOI: 10.1016/j.biortech.2003.12.015
Camassola, M., da Rosa, L. O., Calloni, R., Gaio, T. A., and Dillon, A. J. P. (2013). “Secretion of laccase and manganese peroxidase by Pleurotus strains cultivate in solid-state using Pinus spp. sawdust,” Braz. J. Microbiol. 44(1), 207-213. DOI: 10.1590/S1517-83822013005000006
Cesarino, I., Araújo, P., Júnior, A. P. D., and Mazzafera, P. (2012). “An overview of lignin metabolism and its effect on biomass recalcitrance,” Braz. J. Bot. 35(4),303-311. DOI: 10.1590/S0100-84042012000400003
Chagas, E. P., and Durrant, L. R. (2001). “Decolorization of azo dyes by Phanerochaete chrysosporium and Pleurotus sajorcaju,” Enzyme Microb. Technol. 29(8-9), 473-477. DOI: 10.1016/S0141-0229(01)00405-7
Champagne, P.-P., and Ramsay, J. A. (2005). “Contribution of manganese peroxidase and laccase to dye decoloration by Trametes versicolor,” Appl. Microbiol. Biotechnol. 69,276-285. DOI: 10.1007/s00253-005-1964-8
Christian, V. V., Shrivastava, R., Novotný, C., and Vyas, B. R. M. (2003). “Decolorization of sulfonphthalein dyes by manganese peroxidase activity of the white-rot fungus Phanerochaete chrysosporium,” Folia Microbiol. 48(6),771-774. DOI: 10.1007/BF02931512
Coates, J. (2000). “Interpretation of infrared spectra, A practical approach,” in: Encyclopedia of Analytical Chemistry, R. A. Meyers (ed.), John Wiley & Sons, Hoboken, NJ, pp. 10815-10837. DOI: 10.1002/9780470027318.a5606
Dashtban, M., Schraft, H., Syed, T., A., and Qin, W. (2010). “Fungal biodegradation and enzymatic modification of lignin,” Int. J. Biochem. Mol. Biol. 1(1),36-50.
de la Rubia, T., Linares, A., Pérez, J., Muñoz-Dorado, J., Romera, J., and Martı́nez, J. (2002). “Characterization of manganese-dependent peroxidase isoenzymes from the ligninolytic fungus Phanerochaete flavido-alba,” Res. Microbiol. 153(8), 547-554. DOI: 10.1016/S0923-2508(02)01357-8
de la Rubia, T., Ruiz, E., Pérez, J., and Martínez, J. (2002). “Properties of a laccase produced by Phanerochaete flavido-alba induced by vanillin,” Arch. Microbiol.179, 70-73. DOI: 10.1007/s00203-002-0501-8
dos Santos Bazanella, G. C., de Souza, D. F., Castoldi, R., Oliveira, R. F., Bracht, A., and Peralta, R. M. (2013).“Production of laccase and manganese peroxidase by Pleurotus pulmonarius in solid-state cultures and application in dye decolorization,” Folia Microbiol. 58(6), 641-647. DOI: 10.1007/s12223-013-0253-7
D’Souza, T. M., Merritt, C. S., and Reddy, C. A. (1999). “Lignin-modifying enzymes of the white rot basidiomycete Ganoderma lucidum,” Appl. Environ. Microbiol. 65(12), 5307-5313.
Eichlerová, I., Homolka, L., Lisá, L., and Nerud, F. (2005). “Orange G and Remazol brilliant blue R decolorization by white rot fungi Dichomitus squalens, Ischnoderma resinosum and Pleurotus calyptratus,” Chemosphere 60(3), 398-404. DOI: 10.1016/j.chemosphere.2004.12.036
Erden, E., Ucar, M. C., Gezer T., and Pazarlioglu, N. K. (2009). “Screening for ligninolytic enzymes from autochthonous fungi and applications for decolorization of Remazol marine blue,” Braz. J. Microbiol. 40(2), 346-353. DOI: 10.1590/S1517-838220090002000026
Erkurt, E. A., Ünyayar, A., and Kumbur, H. (2007). “Decolorization of synthetic dyes by white rot fungi, involving laccase enzyme in the process,” Process Biochem. 42(10), 1429-1435. DOI: 10.1016/j.procbio.2007.07.011
Felsenstein, J. (1985). “Confidence limits on phylogenies: An approach using the bootstrap,” Evolution 39(4), 783-791. DOI: 10.1111/j.1558-5646.1985.tb00420.x
Glenn, J. K., and Gold, M. H. (1985). “Purification and characterization of an extracellular Mn(II)-dependent peroxidase from the lignin-degrading basidiomycete, Phanerochaete chrysosporium,” Arch. Biochem. Biophys. 242(2), 329-341.
Harazono, K., Watanabe, Y., and Nakamura, K. (2003). “Decolorization of azo dye by the white-rot basidiomycete Phanerochaete sordida and by its manganese peroxidase,” J. Biosci. Bioeng. 95(5), 455-495. DOI:10.1016/S1389-1723(03)80044-0
Hildén, K., Bortfeldt, R., Hofrichter, M., Hatakka, A., and Lundell, T. (2008). “Molecular characterization of the basidiomycete isolate Nematoloma frowardii b19 and its manganese peroxidase places the fungus in the corticioid genus Phlebia,” Microbiol.154, 2371-2379. DOI: 10.1099/mic.0.2008/018747-0
Hofrichter, M. (2002). “Review: Lignin conversion by manganese peroxidase (MnP),” Enzyme Microb. Technol. 30(4), 454-466. DOI:10.1016/S0141-0229(01)00528-2
Hoshino, F., Kajino, T., Sugiyama, H., Asami, O., and Takahashi, H. (2002). “Thermally stable and hydrogen peroxide tolerant manganese peroxidase (MnP) from Lenzites betulinus,” FEBS Lett. 530(1-3), 249-252. DOI:10.1016/S0014-5793(02)03454-3
Ire, F. S., and Ahuekwe, E. F. (2016). “Production of fungal laccase using orange peelings as substrate by submerged static fermentation,” Br. Microbiol. Res. J. 15(5), 1-19. DOI:10.9734/BMRJ/2016/27257
Jarosz-Wilkołazka, A., Luterek, J., and Olszewska, A. (2009). “Catalytic activity of versatile peroxidase from Bjerkandera fumosa at different pH,” Biocatal. Biotransform. 26(4), 280-287.DOI: 10.1080/10242420701830082
Järvinen, J., Taskila, S., Isomäki, R., and Ojamo, H. (2012). “Screening of white-rot fungi manganese peroxidases: A comparison between the specific activities of the enzyme from different native producers,” AMB Express 2(1), 62-70. DOI:10.1186/2191-0855-2-62
Kalkan, E., Nadaroğlu, H., Celebi, N., and Tozsin, G. (2014). “Removal of textile dye Reactive Black 5 from aqueous solution by adsorption on laccase-modified silica fume,” Desalin. Water Treat. 52(31-33), 6122-6134. DOI:10.1080/19443994.2013.811114
Kong, W., Chen, H., Lyu, S., Ma, F., Yu, H., and Zhang, X. (2016). “Characterization of a novel manganese peroxidase from white-rot fungus Echinodontium taxodii 2538, and its use for the degradation of lignin-related compounds,” Process Biochem. 51(11), 1776-1783. DOI: 10.1016/j.procbio.2016.01.007
Kinnunen, A., Maijala, P., Järvinen, P., and Hatakka, A. (2016). “Improved efficiency in screening for lignin-modifying peroxidases and laccases of basidiomycetes,” Curr. Biotechnol. 6(2), 105-115. DOI: 10.2174/2211550105666160330205138
Kiiskinen, L.-L., Rättö, M., and Kruus, K. (2004). “Screening for novel laccase-producing microbes,” J. Appl. Microbiol. 97(3), 640-646. DOI: 10.1111/j.1365-2672.2004.02348.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). “MEGA X: Molecular evolutionary genetics analysis across computing platforms,” Mol. Biol. Evol. 35(6), 1547-1549. DOI: 10.1093/molbev/msy096
Lankinen, P. V., Bonnen, A. M., Anton, L. H., Wood, D. A., Kalkkinen, N., Hatakka, A., and Thurston, C. F.(2001). “Characteristics and N-terminal amino acid sequence of manganese peroxidase from solid substrate cultures of Agaricus bisporus,” Appl. Microbiol. Biotechnol. 55(2), 170-176.
Lisov, A. V., Leontievsky, A. A., and Golovleva, L. A. (2003). “Hybrid Mn-peroxidase from the ligninolytic fungus Panus tigrinus 8/18. Isolation, substrate specificity, and catalytic cycle,” Biochemistry (Moscow) 68(9), 1027-1035. DOI: 10.1023/A:1026072815106
Lu, Y., Lu, Y.-C., Hu, H.-Q., Xie, F.-J., Wei, X.-Y., and Fan, X. (2017). “Structural characterization of lignin and its degradation products with spectroscopic methods,” J. Spectrosc. 2017, article ID 8951658. DOI:10.1155/2017/8951658
Madadi, M., and Abbas, A. (2017). “Lignin degradation by fungal pretreatment: A review,” J. Plant Pathol. Microbiol. 8(2), 1-6. DOI:10.4172/2157-7471.1000398
Maltseva, O.V., Niku-Paavola, M. L., Leontievsky, A. A., Myasoedova, N. M., and Golovleva, L. A. (1991). “Ligninolytic enzymes of the white-rot fungus Panus tigrinus,” Biotechnol. Appl. Biochem. 13(3), 291-302.
Mester, T., and Field, J. A. (1997). “Optimization of manganese peroxidase production by the white rot fungus Bjerkandera sp. strain BOS55,” FEMS Microbiol. Lett.155(2), 161-168. DOI:10.1111/j.1574-6968.1997.tb13873.x
Murugesan, K., and Kalaichelvan, P. T. (2003). “Synthetic dye decolourization by white rot fungi,” Indian J. Exp. Biol. 41(9), 1076-1087.
Novotný, C., Svobodová, K., Erbanová, P., Cajthaml, T., Kasinath, A., Lang, E., and Šašek, V. (2004). “Lignolytic fungi in bioremediation: Extracellular enzyme production and degradation rate,” Soil Biol. Biochem. 36(10), 1545-1551. DOI: 10.1016/j.soilbio.2004.07.019
Palma, C., Martínez, A. T., Lema, J. M., and Martínez, M. J. (2000). “Different fungal manganese-oxidizing peroxidases: A comparison between Bjerkandera sp. and Phanerochaete chrysosporium,” J. Biotechnol. 77(2-3), 235-245.
Palmieri, G., Cennamo, G., and Sannia, G. (2005). “Remazol brilliant blue R decolorization by the fungus Pleurotus ostreatus and its oxidative enzymatic system,” Enzyme Microb. Technol. 36, 17-24.
Patrick, F., Mtui, G., Mshandete, A. M., and Kivaisi, A. (2011). “Optimization of laccase and manganese peroxidase production in submerged culture of Pleurotus sajorcaju,” Afr. J. Biotechnol. 10(50), 10166-10177. DOI: 10.5897/AJB11.863
Perez, J., and Jeffries, T. W. (1992). “Roles of manganese and organic acid chelators in regulating lignin degradation and biosynthesis of peroxidases by Phanerochaete chrysosporium,” Appl. Environ. Microbiol. 58(8), 2402-2409.
Pointing, S. B. (2001). “Feasibility of bioremediation by white-rot fungi,” Appl. Microbiol. Biotechnol. 57(1-2), 20-33. DOI: 10.1007/s002530100745
Pollegioni, L., Tonin, F., and Rosini, E. (2015). “Lignin-degrading enzymes,” FEBS J. 282(7), 1190-1213. DOI:10.1111/febs.13224
Pradeep, K. V., Chandrashekar, N., and Manpal, S. (2013). “Screening of white rot basidiomycetes for novel laccases,” Adv. Appl. Res. 5(1), 84-89.
Qin, X., Zhang, J., Zhang, X., and Yang, Y. (2014). “Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye,” PLoS ONE 9(11). DOI: 10.1371/journal.pone.0113282
Rao, R. G., Ravichandran, A., Dhali, A., Kolte, A. P., Giridhar, K., and Manpal, S. (2018). “A rapid microwave method for isolation of genomic DNA and identification of white rot fungi,” Biotechnol. J. Int. 21(2), 1-7.DOI: 10.1101/307066
Sahadevan, L. D. M., Misra, C. S., and Thankamani, V. (2016). “Characterization of lignin-degrading enzymes (LDEs) from a dimorphic novel fungus and identification of products of enzymatic breakdown of lignin,” 3 Biotech 6(1), 56-71. DOI: 10.1007/s13205-016-0384-z
Saitou, N., and Nei, M. (1987). “The neighbor-joining method: A new method for reconstructing phylogenetic trees,” Mol. Biol. Evol. 4(4),406-425. DOI: 10.1093/oxfordjournals.molbev.a040454
Shrivastava, R., Christian, V., and Vyas, B. R. M. (2005). “Enzymatic decolorization of sulfonphthalein dyes,” Enzyme Microb. Technol. 36(2-3), 333-337. DOI: 10.1016/j.enzmictec.2004.09.004
Songulashvili, G., Elisashvili, V., Hadar, Y., and Nevo, E. (2008). “Effect of the carbon source and inoculum preparation method on laccase and manganese peroxidase production in submerged cultivation by the medicinal mushroom Ganoderma lucidium (W. Curt.: Fr.) P. Karst. (Aphyllophoromycetideae),” Int. J. Med. Mushrooms 10(1),79-86. DOI: 10.1615/IntJMedMushr.v10.i1.100
Takano, M., Nakamura, M., Nishida, A., and Ishihara, M. (2004). “Manganese peroxidase from Phanerochaete crassa WD1694,” Bulletin of FFPRI 3(1), 7-13.
Tamura, K., Nei, M., and Kumar, S. (2004). “Prospects for inferring very large phylogenies by using the neighbor-joining method,” Proc. Natl. Acad. Sci. U. S. A.101(30), 11030-11035. DOI: 10.1073/pnas.0404206101
Usha, K. Y., Praveen, K., and Reddy, B. R. (2014). “Enhanced production of ligninolytic enzymes by a mushroom Stereum ostrea,” Biotechnol. Res. Int. 2014, 1-9. DOI: 10.1155/2014/815495
Van Aken, B., and Agathos, S. (2002). “Implication of manganese (III), oxalate, and oxygen in the degradation of nitroaromatic compounds by manganese peroxidase (MnP),” Appl. Microbiol. Biotechnol. 58(3), 345-351. DOI:10.1007/s00253-001-0888-1
Vasina, D. V., Moiseenko, K. V., Fedorova, T. V., and Tyazhelova, T. V. (2017). “Lignin-degrading peroxidases in white-rot fungus Trametes hirsuta 072. Absolute expression quantification of full multigene family,” PLoS ONE 12(3). DOI: 10.1371/journal.pone.0173813
Wang, X., Yao, B., and Su, X. (2018). “Linking enzymatic oxidative degradation of lignin to organics detoxification,” Int. J. Mol. Sci. 19(11), 3373-3389. DOI: 10.3390/ijms19113373
Wariishi, H., Valli, K., and Gold, M. (1992). “Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators,” J. Biol. Chem. 267(33), 23688-23695.
Wesenberg, D., Kyriakides, I., and Agathos, S. N. (2003). “White-rot fungi and their enzymes for the treatment of industrial dye effluents,” Biotechnol. Adv. 22(1-2), 161-187. DOI: 10.1016/j.biotechadv.2003.08.011
Yang, X., Ma, F., Zeng, Y., Yu, H., Xu, C., and Zhang, X. (2010). “Structure alteration of lignin in corn stover degraded by white-rot fungus Irpex lacteus CD2,” Int. Biodeterior. Biodegrad. 64(2), 119-123. DOI:10.1016/j.ibiod.2009.12.001
Zhang, H., Zhang, J., Zhang, X., and Geng, A. (2018). “Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching,” Process Biochem. 66, 222-229. DOI: 10.1016/j.procbio.2017.12.011
Zhao, X., Huang, X., Yao, J., Zhou, Y., and Jia, R. (2015). “Fungal growth and manganese peroxidase production in a deep tray solid-state bioreactor, and in vitro decolorization of poly R-478 by MnP,” J. Microbiol. Biotechnol. 25(6), 803-813. DOI: 10.4014/jmb.1410.10054
Article submitted: May 5, 2019; Peer review completed: June 17, 2019; Revised version received and accepted: June 20, 2019; Published: June 28, 2019.
DOI: 10.15376/biores.14.3.6558-6576
